3ONR
 
 | | Crystal structure of the calcium chelating immunodominant antigen, calcium dodecin (Rv0379),from Mycobacterium tuberculosis with a novel calcium-binding site | | Descriptor: | CALCIUM ION, FORMIC ACID, PLATINUM (II) ION, ... | | Authors: | Arulandu, A, Sacchettini, J.C. | | Deposit date: | 2010-08-30 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of calcium dodecin (Rv0379), from Mycobacterium tuberculosis with a unique calcium-binding site.
Protein Sci., 20, 2011
|
|
2VAE
 
 | | Fast maturing red fluorescent protein, DsRed.T4 | | Descriptor: | 1,2-ETHANEDIOL, RED FLUORESCENT PROTEIN | | Authors: | Strongin, D.E, Bevis, B, Khuong, N, Downing, M.E, Strack, R.L, Sundaram, K, Glick, B.S, Keenan, R.J. | | Deposit date: | 2007-08-31 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Rearrangements Near the Chromophore Influence the Maturation Speed and Brightness of Dsred Variants.
Protein Eng.Des.Sel., 20, 2007
|
|
2VAX
 
 | | Crystal structure of deacetylcephalosporin C acetyltransferase (Cephalosporin C-soak) | | Descriptor: | 4-(3-ACETOXYMETHYL-2-CARBOXY-8-OXO-5-THIA-1-AZA-BICYCLO[4.2.0]OCT-2-EN-7-YLCARBAMOYL)-1-CARBOXY-BUTYL-AMMONIUM, ACETATE ION, ACETYL-COA--DEACETYLCEPHALOSPORIN C ACETYLTRANSFERASE | | Authors: | Lejon, S, Ellis, J, Valegard, K. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Last Step in Cephalosporin C Formation Revealed: Crystal Structures of Deacetylcephalosporin C Acetyltransferase from Acremonium Chrysogenum in Complexes with Reaction Intermediates.
J.Mol.Biol., 377, 2008
|
|
3HBX
 
 | | Crystal structure of GAD1 from Arabidopsis thaliana | | Descriptor: | Glutamate decarboxylase 1 | | Authors: | Gut, H, Dominici, P, Pilati, S, Gruetter, M.G, Capitani, G. | | Deposit date: | 2009-05-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.672 Å) | | Cite: | A common structural basis for pH- and calmodulin-mediated regulation in plant glutamate decarboxylase.
J.Mol.Biol., 392, 2009
|
|
3H6O
 
 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-(2-fluorobenzyl)-2,4-dimethyl-4,6-dihydro-5H-thieno[2',3':4,5]pyrrolo[2,3-d]pyridazin-5-one, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-23 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activator-Bound Structures of Human Pyruvate Kinase M2
to be published
|
|
3HB3
 
 | | High resolution crystal structure of Paracoccus denitrificans cytochrome c oxidase | | Descriptor: | ANTIBODY FV FRAGMENT, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Koepke, J, Angerer, H, Peng, G. | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High resolution crystal structure of Paracoccus denitrificans cytochrome c oxidase: New insights into the active site and the proton transfer pathways
Biochim.Biophys.Acta, 1787, 2009
|
|
2VC7
 
 | | Structural basis for natural lactonase and promiscuous phosphotriesterase activities | | Descriptor: | (4S)-4-(decanoylamino)-5-hydroxy-3,4-dihydro-2H-thiophenium, 1,2-ETHANEDIOL, ARYLDIALKYLPHOSPHATASE, ... | | Authors: | Elias, M, Dupuy, J, Merone, L, Mandrich, L, Moniot, S, Rochu, D, Lecomte, C, Rossi, M, Masson, P, Manco, G, Chabriere, E. | | Deposit date: | 2007-09-19 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Natural Lactonase and Promiscuous Phosphotriesterase Activities.
J.Mol.Biol., 379, 2008
|
|
3BLZ
 
 | |
3VC2
 
 | | Crystal structure of geranyl diphosphate C-methyltransferase from Streptomyces coelicolor A3(2) in complex with Mg2+, geranyl diphosphate, and S-adenosyl-L-homocysteine | | Descriptor: | GERANYL DIPHOSPHATE, Geranyl diphosphate 2-C-methyltransferase, MAGNESIUM ION, ... | | Authors: | Koksal, M, Christianson, D.W. | | Deposit date: | 2012-01-03 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Structure of Geranyl Diphosphate C-Methyltransferase from Streptomyces coelicolor and Implications for the Mechanism of Isoprenoid Modification.
Biochemistry, 51, 2012
|
|
3HBB
 
 | | Structures of dihydrofolate reductase-thymidylate synthase of Trypanosoma cruzi in the folate-free state and in complex with two antifolate drugs, trimetrexate and methotrexate | | Descriptor: | 1,2-ETHANEDIOL, Dihydrofolate reductase-thymidylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of dihydrofolate reductase-thymidylate synthase of Trypanosoma cruzi in the folate-free state and in complex with two antifolate drugs, trimetrexate and methotrexate.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2VIW
 
 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | 4-(2-aminoethoxy)-N-(3-chloro-2-ethoxy-5-piperidin-1-ylphenyl)-3,5-dimethylbenzamide, ACETATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR CHAIN B | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator.
J.Med.Chem., 51, 2008
|
|
2VAV
 
 | | Crystal structure of deacetylcephalosporin C acetyltransferase (DAC-Soak) | | Descriptor: | 4-(3-ACETOXYMETHYL-2-CARBOXY-8-OXO-5-THIA-1-AZA-BICYCLO[4.2.0]OCT-2-EN-7-YLCARBAMOYL)-1-CARBOXY-BUTYL-AMMONIUM, ACETATE ION, ACETYL-COA--DEACETYLCEPHALOSPORIN C ACETYLTRANSFERASE | | Authors: | Lejon, S, Ellis, J, Valegard, K. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-23 | | Last modified: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The last step in cephalosporin C formation revealed: crystal structures of deacetylcephalosporin C acetyltransferase from Acremonium chrysogenum in complexes with reaction intermediates.
J. Mol. Biol., 377, 2008
|
|
2VIP
 
 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | 4-(2-AMINOETHOXY)-3,5-DICHLORO-N-[3-(1-METHYLETHOXY)PHENYL]BENZAMIDE, ACETATE ION, SULFATE ION, ... | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator
J.Med.Chem., 51, 2008
|
|
3CW1
 
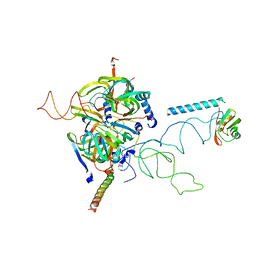 | | Crystal Structure of Human Spliceosomal U1 snRNP | | Descriptor: | Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, Small nuclear ribonucleoprotein G, ... | | Authors: | Pomeranz Krummel, D.A, Oubridge, C, Leung, A.K, Li, J, Nagai, K. | | Deposit date: | 2008-04-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.493 Å) | | Cite: | Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution.
Nature, 458, 2009
|
|
2VC8
 
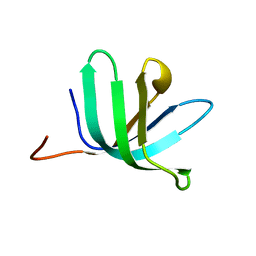 | |
2V9P
 
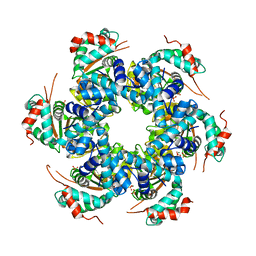 | | Crystal structure of papillomavirus E1 hexameric helicase DNA-free form | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, REPLICATION PROTEIN E1 | | Authors: | Sanders, C.M, Kovalevskiy, O.V, Sizov, D, Lebedev, A.A, Isupov, M.N, Antson, A.A. | | Deposit date: | 2007-08-24 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Papillomavirus E1 Helicase Assembly Maintains an Asymmetric State in the Absence of DNA and Nucleotide Cofactors.
Nucleic Acids Res., 35, 2007
|
|
3VIC
 
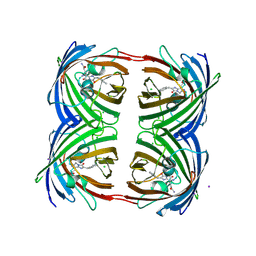 | | Green-fluorescent variant of the non-fluorescent chromoprotein Rtms5 | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Traore, D.A.K, Byres, E, Wilce, M, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2011-09-28 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Green Fluorescent Protein Containing a QFG Tri-Peptide Chromophore: Optical Properties and X-Ray Crystal Structure.
Plos One, 7, 2012
|
|
3QQM
 
 | |
3Q7H
 
 | | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, DI(HYDROXYETHYL)ETHER | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii
To be Published
|
|
3D4G
 
 | |
2VIV
 
 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | 4-(2-aminoethoxy)-N-(3-chloro-5-piperidin-1-ylphenyl)-3,5-dimethylbenzamide, ACETATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR CHAIN B | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator.
J.Med.Chem., 51, 2008
|
|
2V3M
 
 | | Structure of the Gar1 domain of NAf1 | | Descriptor: | NAF1, SULFATE ION | | Authors: | Leulliot, N, Godin, K.S, Hoareau-Aveilla, C, Quevillon-Cheruel, S, Varani, G, Henry, Y, van Tilbeurgh, H. | | Deposit date: | 2007-06-19 | | Release date: | 2007-07-10 | | Last modified: | 2011-08-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The Box H/Aca Rnp Assembly Factor Naf1P Contains a Domain Homologous to Gar1P Mediating its Interaction with Cbf5P.
J.Mol.Biol., 371, 2007
|
|
3QJG
 
 | | Epidermin biosynthesis protein EpiD from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, Epidermin biosynthesis protein EpiD, FLAVIN MONONUCLEOTIDE | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-28 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Epidermin biosynthesis protein EpiD from Staphylococcus aureus.
To be Published
|
|
3D29
 
 | | Proteasome Inhibition by Fellutamide B | | Descriptor: | (3R)-3-HYDROXYDODECANOIC ACID, Fellutamide B, PRE10 isoform 1, ... | | Authors: | Groll, M, Hines, J, Fahnestock, M, Crews, M.C. | | Deposit date: | 2008-05-07 | | Release date: | 2008-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Proteasome Inhibition by Fellutamide B Induces Nerve Growth Factor Synthesis
Chem.Biol., 15, 2008
|
|
2VAT
 
 | | Crystal structure of deacetylcephalosporin C acetyltransferase in complex with coenzyme A | | Descriptor: | ACETATE ION, ACETYL-COA--DEACETYLCEPHALOSPORIN C ACETYLTRANSFERASE, COENZYME A, ... | | Authors: | Lejon, S, Ellis, J, Valegard, K. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Last Step in Cephalosporin C Formation Revealed: Crystal Structures of Deacetylcephalosporin C Acetyltransferase from Acremonium Chrysogenum in Complexes with Reaction Intermediates.
J.Mol.Biol., 377, 2008
|
|
