6G6O
 
 | | Crystal structure of the computationally designed Ika8 protein: crystal packing No.1 in P63 | | Descriptor: | GLYCEROL, Ika8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
8JH6
 
 | | Crystal structure of apixaban-bound racemic ABLE | | Descriptor: | 1-(4-METHOXYPHENYL)-7-OXO-6-[4-(2-OXOPIPERIDIN-1-YL)PHENYL]-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDINE-3-CARBOXAMIDE, D-ABLE, L-ABLE | | Authors: | Zhao, K, Li, T. | | Deposit date: | 2023-05-22 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of apixaban-bound racemic ABLE
To Be Published
|
|
7LZ3
 
 | | Computational design of constitutively active cGAS | | Descriptor: | Cyclic GMP-AMP synthase, GLYCEROL, ZINC ION | | Authors: | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8JYT
 
 | | Ancestral imine reducatase N560 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ancestra imine reductase | | Authors: | Zhu, X.X. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The structure of ancestral imine reductase N560.
To Be Published
|
|
6H7I
 
 | |
6UGC
 
 | | C3 symmetric peptide design number 3 | | Descriptor: | C3-3 cyclic peptide design, CADMIUM ION, SODIUM ION | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6G6N
 
 | | Crystal structure of the computationally designed Tako8 protein in C2 | | Descriptor: | Tako8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
6FPA
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_G01 in space group P212121 | | Descriptor: | DARPin 1238_G01 | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6I3B
 
 | |
8JKU
 
 | | Ancestal imine reductase | | Descriptor: | Ancestral imine reductase N559, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhu, X.X, Zheng, G.W. | | Deposit date: | 2023-06-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of ancestral imine reductase at 2.56 Angstroms resolution.
To Be Published
|
|
6I37
 
 | | Crystal structure of nv1Pizza6-AYW, a circularly permuted designer protein | | Descriptor: | SULFATE ION, nv1Pizza6-AYW | | Authors: | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | Deposit date: | 2018-11-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
6I39
 
 | | Crystal structure of v31Pizza6-AYW, a circularly permuted designer protein | | Descriptor: | MAGNESIUM ION, v31Pizza6-AYW | | Authors: | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | Deposit date: | 2018-11-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
8OT7
 
 | |
6I1J
 
 | |
6I38
 
 | |
6IMG
 
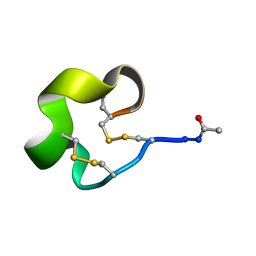 | | Solution Structure of Bicyclic Peptide pb-13 | | Descriptor: | (ACE)-GLY-CYS-PRO-CYS-ILE-TRP-PRO-GLU-LEU-CYS-PRO-TRP-ILE-ARG-SER-CYS-(NH2) | | Authors: | Yao, H, Lin, P, Zha, J, Zha, M, Zhao, Y, Wu, C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Method: | SOLUTION NMR | | Cite: | Ordered and Isomerically Stable Bicyclic Peptide Scaffolds Constrained through Cystine Bridges and Proline Turns.
Chembiochem, 20, 2019
|
|
6I3A
 
 | | Crystal structure of v22Pizza6-AYW, a circularly permuted designer protein | | Descriptor: | BROMIDE ION, v22Pizza6-AYW | | Authors: | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | Deposit date: | 2018-11-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
8ATC
 
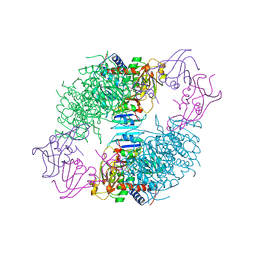 | | COMPLEX OF N-PHOSPHONACETYL-L-ASPARTATE WITH ASPARTATE CARBAMOYLTRANSFERASE. X-RAY REFINEMENT, ANALYSIS OF CONFORMATIONAL CHANGES AND CATALYTIC AND ALLOSTERIC MECHANISMS | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE (R STATE), CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ... | | Authors: | Ke, H, Lipscomb, W.N, Cho, Y, Honzatko, R.B. | | Deposit date: | 1989-08-25 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complex of N-phosphonacetyl-L-aspartate with aspartate carbamoyltransferase. X-ray refinement, analysis of conformational changes and catalytic and allosteric mechanisms.
J.Mol.Biol., 204, 1988
|
|
8AED
 
 | |
8BAO
 
 | |
8B9Y
 
 | | Cysteine Synthase from Trypanosoma cruzi with PLP and OAS | | Descriptor: | Cysteine synthase, putative, GLYCEROL, ... | | Authors: | Sowerby, K.V, Pohl, E. | | Deposit date: | 2022-10-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cysteine synthase: multiple structures of a key enzyme in cysteine synthesis and a potential drug target for Chagas disease and leishmaniasis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B9W
 
 | | Cysteine Synthase from Trypanosoma theileri with PLP bound | | Descriptor: | CHLORIDE ION, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Sowerby, K.V, Pohl, E. | | Deposit date: | 2022-10-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Cysteine synthase: multiple structures of a key enzyme in cysteine synthesis and a potential drug target for Chagas disease and leishmaniasis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B9M
 
 | | Cysteine Synthase from Leishmania Infantum | | Descriptor: | CHLORIDE ION, Cysteine synthase, GLYCEROL | | Authors: | Sowerby, K.V, Pohl, E. | | Deposit date: | 2022-10-06 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cysteine synthase: multiple structures of a key enzyme in cysteine synthesis and a potential drug target for Chagas disease and leishmaniasis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BGJ
 
 | |
8B45
 
 | | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4 | | Descriptor: | 1,2-ETHANEDIOL, CC-Tri-(UbUc)4, SODIUM ION, ... | | Authors: | Kumar, P, Martin, F.J.O, Dawson, W.M, Zieleniewski, F, Woolfson, D.N. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4
To Be Published
|
|
