1EI2
 
 | | STRUCTURAL BASIS FOR RECOGNITION OF THE RNA MAJOR GROOVE IN THE TAU EXON 10 SPLICING REGULATORY ELEMENT BY AMINOGLYCOSIDE ANTIBIOTICS | | Descriptor: | NEOMYCIN, TAU EXON 10 SRE RNA | | Authors: | Varani, L, Spillantini, M.G, Goedert, M, Varani, G. | | Deposit date: | 2000-02-23 | | Release date: | 2000-03-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of the RNA major groove in the tau exon 10 splicing regulatory element by aminoglycoside antibiotics.
Nucleic Acids Res., 28, 2000
|
|
469D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
247D
 
 | | CRYSTAL STRUCTURES OF AN A-FORM DUPLEX WITH SINGLE-ADENOSINE BULGES AND A CONFORMATIONAL BASIS FOR SITE SPECIFIC RNA SELF-CLEAVAGE | | Descriptor: | DNA/RNA (5'-R(*GP*CP*GP*)-D(*AP*TP*AP*TP*AP*)-R(*CP*GP*C)-3') | | Authors: | Portmann, S, Grimm, S, Workman, C, Usman, N, Egli, M. | | Deposit date: | 1996-02-02 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of an A-form duplex with single-adenosine bulges and a conformational basis for site-specific RNA self-cleavage.
Chem.Biol., 3, 1996
|
|
8DGA
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IV | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DG7
 
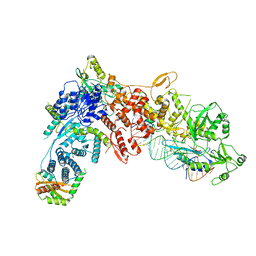 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex III | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DFV
 
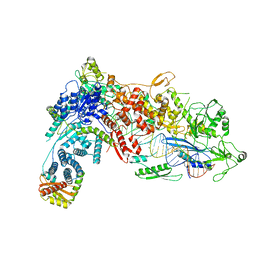 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IIa | | Descriptor: | CALCIUM ION, Endoribonuclease Dcr-1, Loquacious, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-22 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DG5
 
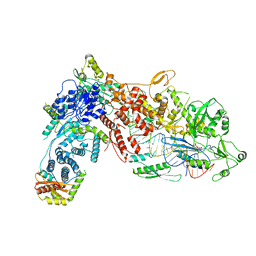 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IIb | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
1ZIG
 
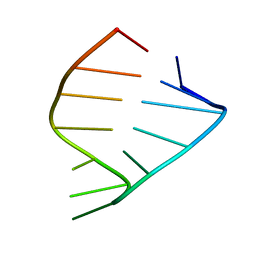 | | GAGA RNA TETRALOOP, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*GP*AP*GP*AP*GP*CP*CP*U)-3') | | Authors: | Jucker, F.M, Heus, H.A, Yip, P.F, Moors, E, Pardi, A. | | Deposit date: | 1996-07-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A network of heterogeneous hydrogen bonds in GNRA tetraloops.
J.Mol.Biol., 264, 1996
|
|
1ZIH
 
 | | GCAA RNA TETRALOOP, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*GP*CP*AP*AP*GP*CP*CP*U)-3') | | Authors: | Jucker, F.M, Heus, H.A, Yip, P.F, Moors, E, Pardi, A. | | Deposit date: | 1996-07-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A network of heterogeneous hydrogen bonds in GNRA tetraloops.
J.Mol.Biol., 264, 1996
|
|
2PCW
 
 | |
1ZIF
 
 | | GAAA RNA TETRALOOP, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*GP*AP*AP*AP*GP*CP*CP*U)-3') | | Authors: | Jucker, F.M, Heus, H.A, Yip, P.F, Moors, E, Pardi, A. | | Deposit date: | 1996-07-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A network of heterogeneous hydrogen bonds in GNRA tetraloops.
J.Mol.Biol., 264, 1996
|
|
1FIX
 
 | |
2MKK
 
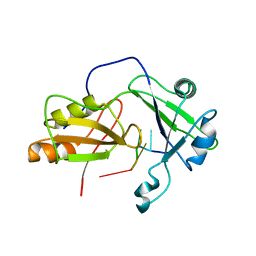 | | Structural model of tandem RRM domains of cytoplasmic polyadenylation element binding protein 1 (CPEB1) in complex with RNA | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 1, RNA (5'-R(*UP*UP*UP*UP*A)-3') | | Authors: | Afroz, T, Skrisovska, L, Belloc, E, Boixet, J.G, Mendez, R, Allain, F.H.-T. | | Deposit date: | 2014-02-07 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A fly trap mechanism provides sequence-specific RNA recognition by CPEB proteins
Genes Dev., 28, 2014
|
|
1G70
 
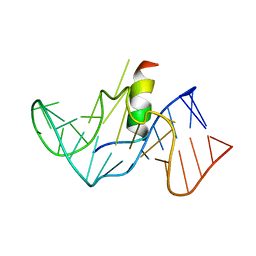 | | COMPLEX OF HIV-1 RRE-IIB RNA WITH RSG-1.2 PEPTIDE | | Descriptor: | HIV-1 RRE-IIB 32 NUCLEOTIDE RNA, RSG-1.2 PEPTIDE | | Authors: | Gosser, Y, Hermann, T, Majumdar, A, Hu, W, Frederick, R, Jiang, F, Xu, W, Patel, D.J. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Peptide-triggered conformational switch in HIV-1 RRE RNA complexes.
Nat.Struct.Biol., 8, 2001
|
|
1YMO
 
 | |
4AL5
 
 | |
7R6Y
 
 | |
3MW6
 
 | | Crystal structure of NMB1681 from Neisseria meningitidis MC58, a FinO-like RNA chaperone | | Descriptor: | GLYCEROL, uncharacterized protein NMB1681 | | Authors: | Tan, K, Zhou, M, Duggan, E, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | N. meningitidis 1681 is a member of the FinO family of RNA chaperones.
Rna Biol., 7, 2010
|
|
3X1L
 
 | |
4TXD
 
 | | Crystal structure of Thermofilum pendens Csc2 | | Descriptor: | Csc2, ZINC ION | | Authors: | Hrle, A, Conti, E. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Structural analyses of the CRISPR protein Csc2 reveal the RNA-binding interface of the type I-D Cas7 family.
Rna Biol., 11, 2014
|
|
1S2M
 
 | |
2QUX
 
 | | PP7 coat protein dimer in complex with RNA hairpin | | Descriptor: | Coat protein, GLYCEROL, RNA (25-MER) | | Authors: | Chao, J.A. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural basis for the coevolution of a viral RNA-protein complex.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2QTX
 
 | | Crystal structure of an Hfq-like protein from Methanococcus jannaschii | | Descriptor: | Uncharacterized protein MJ1435 | | Authors: | Nielsen, J.S, Boggild, A, Andersen, C.B.F, Nielsen, G, Boysen, A, Brodersen, D.E, Valentin-Hansen, P. | | Deposit date: | 2007-08-03 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Hfq-like protein in archaea: Crystal structure and functional characterization of the Sm protein from Methanococcus jannaschii.
Rna, 13, 2007
|
|
6MN8
 
 | |
2RSK
 
 | | RNA aptamer against prion protein in complex with the partial binding peptide | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3'), partial binding peptide of Major prion protein | | Authors: | Mashima, T, Nishikawa, F, Kamatari, Y.O, Fujiwara, H, Nishikawa, S, Kuwata, K, Katahira, M. | | Deposit date: | 2012-03-08 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Anti-prion activity of an RNA aptamer and its structural basis
Nucleic Acids Res., 41, 2013
|
|
