1ELX
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (S102A) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stec, B, Hehir, M, Brennan, C, Nolte, M, Kantrowitz, E.R. | | Deposit date: | 1998-02-10 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Kinetic and X-ray structural studies of three mutant E. coli alkaline phosphatases: insights into the catalytic mechanism without the nucleophile Ser102.
J.Mol.Biol., 277, 1998
|
|
6Q4K
 
 | | CDK2 in complex with FragLite38 | | Descriptor: | (~{E})-3-[3-[(4-chlorophenyl)carbamoyl]phenyl]prop-2-enoic acid, 1,2-ETHANEDIOL, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
7MXY
 
 | | Cryo-EM structure of PfFNT-inhibitor complex | | Descriptor: | (Z)-4,4,5,5,5-pentakis(fluoranyl)-1-(4-methoxy-2-oxidanyl-phenyl)-3-oxidanyl-pent-2-en-1-one, Formate-nitrite transporter | | Authors: | Yu, E.W, Su, C, Lyu, M. | | Deposit date: | 2021-05-19 | | Release date: | 2021-12-08 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Structural basis of transport and inhibition of the Plasmodium falciparum transporter PfFNT
EMBO Rep, 22, 2021
|
|
8R10
 
 | |
9M1H
 
 | | Cryo-EM structure of PGE2-EP1-Gq complex | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Meng, X, Xu, Y, Xu, H.E. | | Deposit date: | 2025-02-26 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural Insights into the Activation of Human Prostaglandin E2 Receptor EP1 Subtype by Prostaglandin E2
To Be Published
|
|
6RFE
 
 | | Human protein kinase CK2 alpha in complex with 2-cyano-2-propenamide compound 4 | | Descriptor: | (~{E})-~{N}-(5-bromanyl-1,3,4-thiadiazol-2-yl)-2-cyano-3-(3-methoxy-4-oxidanyl-phenyl)prop-2-enamide, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Zanforlin, E, Ribaudo, G, Zagotto, G, Battistutta, R, Lolli, G. | | Deposit date: | 2019-04-13 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A novel class of selective CK2 inhibitors targeting its open hinge conformation.
Eur.J.Med.Chem., 195, 2020
|
|
9J89
 
 | | zbp1 nucleic acid complex | | Descriptor: | DNA (5'-D(P*CP*AP*CP*GP*CP*A)-3'), MAGNESIUM ION, RNA (5'-R(P*UP*GP*CP*GP*UP*G)-3'), ... | | Authors: | Gao, A.M, Zhou, C. | | Deposit date: | 2024-08-20 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | ZBP1 senses spliceosome stress through Z-RNA:DNA hybrid recognition.
Mol.Cell, 85, 2025
|
|
6RCM
 
 | | Human protein kinase CK2 alpha in complex with 2-cyano-2-propenamide compound 3 | | Descriptor: | (~{E})-~{N}-(5-~{tert}-butyl-1,3,4-thiadiazol-2-yl)-2-cyano-3-(3-methoxy-4-oxidanyl-phenyl)prop-2-enamide, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Zanforlin, E, Ribaudo, G, Zagotto, G, Battistutta, R, Lolli, G. | | Deposit date: | 2019-04-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel class of selective CK2 inhibitors targeting its open hinge conformation.
Eur.J.Med.Chem., 195, 2020
|
|
5J4D
 
 | |
6RFF
 
 | | Human protein kinase CK2 alpha in complex with 2-cyano-2-propenamide compound 7 | | Descriptor: | (~{E})-~{N}-(5-bromanyl-1,3,4-thiadiazol-2-yl)-2-cyano-3-(3-nitro-4-oxidanyl-phenyl)prop-2-enamide, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Zanforlin, E, Ribaudo, G, Zagotto, G, Battistutta, R, Lolli, G. | | Deposit date: | 2019-04-14 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel class of selective CK2 inhibitors targeting its open hinge conformation.
Eur.J.Med.Chem., 195, 2020
|
|
9MO0
 
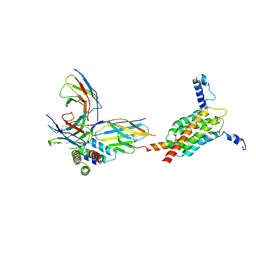 | | Cryo-EM structure of human MPC in complex with AKOS005153046 | | Descriptor: | (~{E})-2-cyano-3-[5-(2-nitrophenyl)furan-2-yl]prop-2-enoic acid, Fab_8D3_2 heavy chain, Fab_8D3_2 light chain, ... | | Authors: | Zhang, J, He, Z, Feng, L. | | Deposit date: | 2024-12-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure of mitochondrial pyruvate carrier and its inhibition mechanism.
Nature, 641, 2025
|
|
9MNX
 
 | | Cryo-EM structure of human MPC in complex with UK5099 in LMNG | | Descriptor: | (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid, Fab_8D3_2 heavy chain, Fab_8D3_2 light chain, ... | | Authors: | Zhang, J, He, Z, Feng, L. | | Deposit date: | 2024-12-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structure of mitochondrial pyruvate carrier and its inhibition mechanism.
Nature, 641, 2025
|
|
9MNZ
 
 | | Cryo-EM structure of human MPC in complex with UK5099 in nanodiscs | | Descriptor: | (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid, Fab_8D3_2 heavy chain, Fab_8D3_2 light chain, ... | | Authors: | Zhang, J, He, Z, Feng, L. | | Deposit date: | 2024-12-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structure of mitochondrial pyruvate carrier and its inhibition mechanism.
Nature, 641, 2025
|
|
6GUR
 
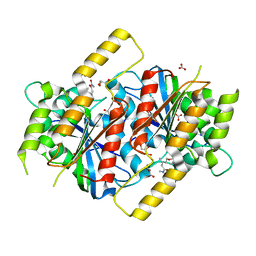 | | Siderophore hydrolase EstB from Aspergillus fumigatus in complex with TAFC | | Descriptor: | (~{Z})-5-[(1~{S},2~{S})-2-acetamido-1-oxidanyl-5-[oxidanyl(propanoyl)amino]pentoxy]-~{N},3-dimethyl-~{N}-oxidanyl-pent-2-enamide, CARBONATE ION, FE (III) ION, ... | | Authors: | Ecker, F, Haas, H, Groll, M, Huber, E.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iron Scavenging in Aspergillus Species: Structural and Biochemical Insights into Fungal Siderophore Esterases.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5KDI
 
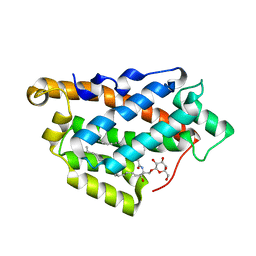 | | How FAPP2 Selects Simple Glycosphingolipids Using the GLTP-fold | | Descriptor: | (~{Z})-~{N}-[(~{E},2~{S},3~{R})-1-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-octadec-4-en-2-yl]octadec-9-enamide, Pleckstrin homology domain-containing family A member 8 | | Authors: | Ochoa-Lizarralde, B, Popov, A.N, Samygina, V.R, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2016-06-08 | | Release date: | 2017-12-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural analyses of 4-phosphate adaptor protein 2 yield mechanistic insights into sphingolipid recognition by the glycolipid transfer protein family.
J.Biol.Chem., 293, 2018
|
|
7GG3
 
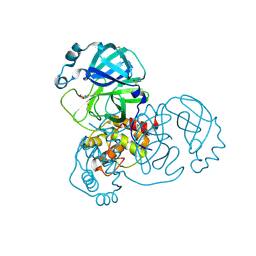 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with AAR-RCN-748c104b-1 (Mpro-x12080) | | Descriptor: | (E)-1-(4,6-dimethoxypyrimidin-2-yl)methanimine, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5KNS
 
 | | E coli hypoxanthine guanine phosphoribosyltransferase in complexed with 9-[(N-phosphonoethyl-N-phosphonoethoxyethyl)-2-aminoethyl]hypoxanthine | | Descriptor: | (2-{[2-(6-oxo-1,6-dihydro-9H-purin-9-yl)ethyl](2-{[(E)-2-phosphonoethenyl]oxy}ethyl)amino}ethyl)phosphonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypoxanthine-guanine phosphoribosyltransferase, ... | | Authors: | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
6UNN
 
 | |
1QOY
 
 | | E.coli Hemolysin E (HlyE, ClyA, SheA) | | Descriptor: | HEMOLYSIN E, SULFATE ION | | Authors: | Wallace, A.J, Stillman, T.J, Atkins, A, Jamieson, S.J, Bullough, P.A, Green, J, Artymiuk, P.J. | | Deposit date: | 1999-11-25 | | Release date: | 2000-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | E. Coli Hemolysin E (Hlye, Clya, Shea): X-Ray Crystal Structure of the Toxin and Observation of Membrane Pores by Electron Microscopy
Cell(Cambridge,Mass.), 100, 2000
|
|
7K5A
 
 | |
1BDX
 
 | | E. COLI DNA HELICASE RUVA WITH BOUND DNA HOLLIDAY JUNCTION, ALPHA CARBONS AND PHOSPHATE ATOMS ONLY | | Descriptor: | DNA (5'-D(P*GP*CP*AP*TP*GP*CP*AP*TP*AP*TP*GP*CP*AP*TP*GP*C)-3'), HOLLIDAY JUNCTION DNA HELICASE RUVA | | Authors: | Hargreaves, D, Rice, D.W, Sedelnikova, S.E, Artymiuk, P.J, Lloyd, R.G, Rafferty, J.B. | | Deposit date: | 1998-05-11 | | Release date: | 1999-11-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal structure of E.coli RuvA with bound DNA Holliday junction at 6 A resolution.
Nat.Struct.Biol., 5, 1998
|
|
7DPS
 
 | |
3IAQ
 
 | | E. coli (lacz) beta-galactosidase (E416V) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Lo, S, Dugdale, M.L, Jeerh, N, Ku, T, Roth, N.J, Huber, R.E. | | Deposit date: | 2009-07-14 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Studies of Glu-416 variants of beta-galactosidase (E. coli) show that the active site Mg(2+) is not important for structure and indicate that the main role of Mg (2+) is to mediate optimization of active site chemistry
Protein J., 29, 2010
|
|
3AAS
 
 | | Bovine beta-trypsin bound to meta-guanidino schiff base copper (II) chelate | | Descriptor: | (E)-N-[(5-carbamimidamido-2-hydroxyphenyl)methylidene]-L-alanine, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Iyaguchi, D, Kawano, S, Toyota, E. | | Deposit date: | 2009-11-26 | | Release date: | 2010-04-07 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the design of novel Schiff base metal chelate inhibitors of trypsin
Bioorg.Med.Chem., 18, 2010
|
|
3GQL
 
 | | Crystal Structure of activated receptor tyrosine kinase in complex with substrates | | Descriptor: | (E)-[4-(3,5-difluorophenyl)-3H-pyrrolo[2,3-b]pyridin-3-ylidene](3-methoxyphenyl)methanol, Basic fibroblast growth factor receptor 1 | | Authors: | Bae, J.H, Lew, E.D, Yuzawa, S, Tome, F, Lax, I, Schlessinger, J. | | Deposit date: | 2009-03-24 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site.
Cell(Cambridge,Mass.), 138, 2009
|
|
