2DPE
 
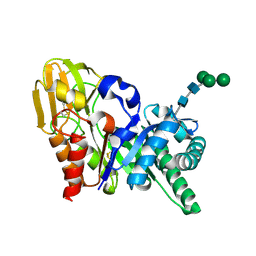 | | Crystal structure of a secretory 40KDA glycoprotein from sheep at 2.0A resolution | | Descriptor: | Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Singh, N, Sharma, S, Das, U, Srinivasan, A, Singh, T.P. | | Deposit date: | 2006-05-11 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of a secretory signalling glycoprotein from sheep at 2.0A resolution
J.Struct.Biol., 156, 2006
|
|
2DPF
 
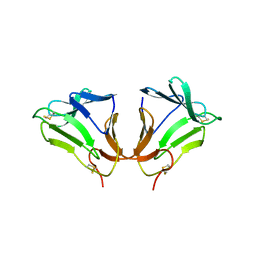 | | Crystal Structure of curculin1 homodimer | | Descriptor: | Curculin, SULFATE ION | | Authors: | Kurimoto, E, Suzuki, M, Amemiya, E, Yamaguchi, Y, Nirasawa, S, Shimba, N, Xu, N, Kashiwagi, T, Kawai, M, Suzuki, E, Kato, K. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Curculin Exhibits Sweet-tasting and Taste-modifying Activities through Its Distinct Molecular Surfaces.
J.Biol.Chem., 282, 2007
|
|
2DPG
 
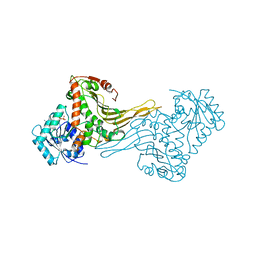 | | COMPLEX OF INACTIVE MUTANT (H240->N) OF GLUCOSE 6-PHOSPHATE DEHYDROGENASE FROM LEUCONOSTOC MESENTEROIDES WITH NADP+ | | Descriptor: | GLUCOSE 6-PHOSPHATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Adams, M.J, Naylor, C.E, Paludin, S, Gover, S. | | Deposit date: | 1998-04-17 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | On the mechanism of the reaction catalyzed by glucose 6-phosphate dehydrogenase.
Biochemistry, 37, 1998
|
|
2DPH
 
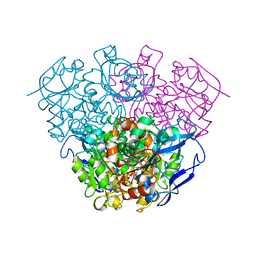 | | Crystal Structure of Formaldehyde dismutase | | Descriptor: | Formaldehyde dismutase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Hasegawa, T, Yamano, A, Yanase, H. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The X-ray crystal structure of formaldehyde dismutase at 2.3 A resolution
Acta Crystallogr.,Sect.A, 58, 2002
|
|
2DPI
 
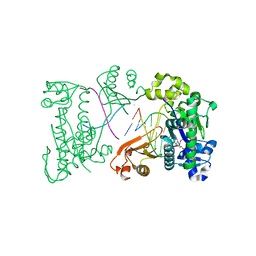 | | Ternary complex of hPoli with DNA and dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*GP*GP*AP*CP*CP*(DOC))-3', 5'-D(*TP*(EDA)P*GP*GP*GP*TP*CP*CP*T)-3', ... | | Authors: | Nair, D.T, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2006-05-12 | | Release date: | 2006-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Hoogsteen base pair formation promotes synthesis opposite the 1,N(6)-ethenodeoxyadenosine lesion by human DNA polymerase iota.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2DPJ
 
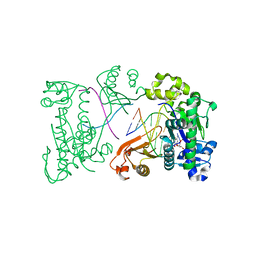 | | structure of hPoli with DNA and dTTP | | Descriptor: | 5'-D(*AP*GP*GP*AP*CP*CP*(DOC))-3', 5'-D(*TP*(EDA)P*GP*GP*GP*TP*CP*CP*T)-3', DNA polymerase iota, ... | | Authors: | Nair, D.T, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2006-05-12 | | Release date: | 2006-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Hoogsteen base pair formation promotes synthesis opposite the 1,N(6)-ethenodeoxyadenosine lesion by human DNA polymerase iota.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2DPK
 
 | |
2DPL
 
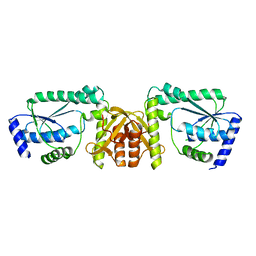 | |
2DPM
 
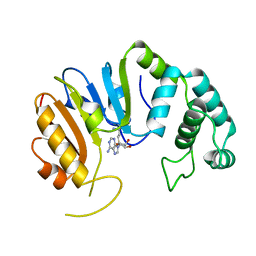 | | DPNM DNA ADENINE METHYLTRANSFERASE FROM STREPTOCCOCUS PNEUMONIAE COMPLEXED WITH S-ADENOSYLMETHIONINE | | Descriptor: | MERCURY (II) ION, PROTEIN (ADENINE-SPECIFIC METHYLTRANSFERASE DPNII 1), S-ADENOSYLMETHIONINE | | Authors: | Tran, P.H, Korszun, Z.R, Cerritelli, S, Springhorn, S.S, Lacks, S.A. | | Deposit date: | 1998-09-03 | | Release date: | 1998-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the DpnM DNA adenine methyltransferase from the DpnII restriction system of streptococcus pneumoniae bound to S-adenosylmethionine.
Structure, 6, 1998
|
|
2DPN
 
 | |
2DPP
 
 | | Crystal structure of thermostable Bacillus sp. RAPc8 nitrile hydratase | | Descriptor: | COBALT (II) ION, Nitrile hydratase alpha subunit, Nitrile hydratase beta subunit | | Authors: | Tsekoa, T.L, Tastan-Bishop, A.O, Cameron, R.A, Sewell, B.T, Sayed, M.F, Cowan, D.A. | | Deposit date: | 2006-05-12 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of thermostable Bacillus sp. RAPc8 nitrile hydratse
To be Published
|
|
2DPQ
 
 | | The crystal structures of the calcium-bound con-G and con-T(K7gamma) dimeric peptides demonstrate a novel metal-dependent helix-forming motif | | Descriptor: | CALCIUM ION, CHLORIDE ION, Conantokin-G | | Authors: | Cnudde, S.E, Prorok, M, Dai, Q, Castellino, F.J, Geiger, J.H. | | Deposit date: | 2006-05-13 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The crystal structures of the calcium-bound con-G and con-T[K7gamma] dimeric peptides demonstrate a metal-dependent helix-forming motif
J.Am.Chem.Soc., 129, 2007
|
|
2DPR
 
 | | The crystal structures of the calcium-bound con-G and con-T(K7Glu) dimeric peptides demonstrate a novel metal-dependent helix-forming motif | | Descriptor: | CALCIUM ION, Conantokin-T | | Authors: | Cnudde, S.E, Prorok, M, Dai, Q, Castellino, F.J, Geiger, J.H. | | Deposit date: | 2006-05-13 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structures of the calcium-bound con-G and con-T[K7gamma] dimeric peptides demonstrate a metal-dependent helix-forming motif
J.Am.Chem.Soc., 129, 2007
|
|
2DPS
 
 | |
2DPT
 
 | |
2DPU
 
 | |
2DPW
 
 | |
2DPX
 
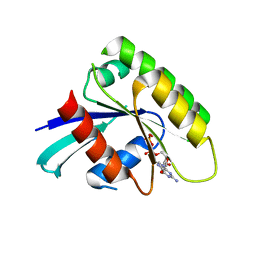 | | Crystal Structure of human Rad GTPase | | Descriptor: | GTP-binding protein RAD, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Yanuar, A, Sakurai, S, Kitano, K, Hakoshima, T. | | Deposit date: | 2006-05-18 | | Release date: | 2006-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Rad GTPase of the RGK-family
Genes Cells, 11, 2006
|
|
2DPY
 
 | |
2DPZ
 
 | |
2DQ0
 
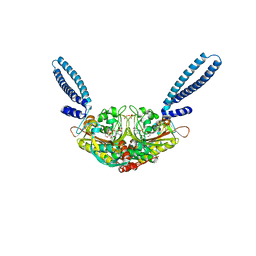 | | Crystal structure of seryl-tRNA synthetase from Pyrococcus horikoshii complexed with a seryl-adenylate analog | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, SULFATE ION, Seryl-tRNA synthetase | | Authors: | Itoh, Y, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-18 | | Release date: | 2007-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic and mutational studies of seryl-tRNA synthetase from the archaeon Pyrococcus horikoshii.
Rna Biol., 5, 2008
|
|
2DQ3
 
 | |
2DQ4
 
 | | Crystal structure of threonine 3-dehydrogenase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, L-threonine 3-dehydrogenase, ZINC ION | | Authors: | Omi, R, Yao, T, Goto, M, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-19 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of threonine 3-dehydrogenase
To be Published
|
|
2DQ5
 
 | | solution structure of the Mid1 B Box2 Chc(D/C)C2H2 Zinc-Binding Domain: insights into an evolutionary conserved ring fold | | Descriptor: | Midline-1, ZINC ION | | Authors: | Massiah, M.A, Matts, J.A.B, Short, K.M, Simmons, B.N, Singireddy, S, Zou, J, Cox, T.C. | | Deposit date: | 2006-05-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the MID1 B-box2 CHC(D/C)C(2)H(2) Zinc-binding Domain: Insights into an Evolutionarily Conserved RING Fold
J.Mol.Biol., 369, 2007
|
|
2DQ6
 
 | | Crystal Structure of Aminopeptidase N from Escherichia coli | | Descriptor: | Aminopeptidase N, SULFATE ION, ZINC ION | | Authors: | Nakajima, Y, Onohara, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2006-05-22 | | Release date: | 2006-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
