1BFF
 
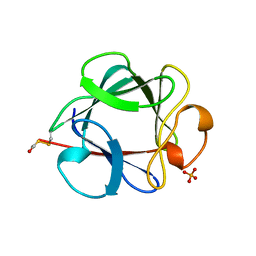 | | THE 154 AMINO ACID FORM OF HUMAN BASIC FIBROBLAST GROWTH FACTOR | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR, BETA-MERCAPTOETHANOL, PHOSPHATE ION | | Authors: | Kastrup, J.S, Eriksson, E.S. | | Deposit date: | 1996-12-06 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of the 154-amino-acid form of recombinant human basic fibroblast growth factor. comparison with the truncated 146-amino-acid form.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1J7T
 
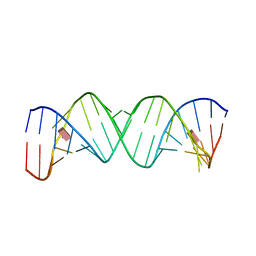 | |
1J7Y
 
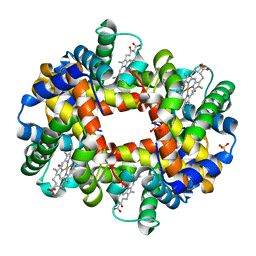 | | Crystal structure of partially ligated mutant of HbA | | Descriptor: | CARBON MONOXIDE, Hemoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miele, A.E, Draghi, F, Arcovito, A, Bellelli, A, Brunori, M, Travaglini-Allocatelli, C, Vallone, B. | | Deposit date: | 2001-05-19 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Control of heme reactivity by diffusion: structural basis and functional characterization in hemoglobin mutants.
Biochemistry, 40, 2001
|
|
2ZEH
 
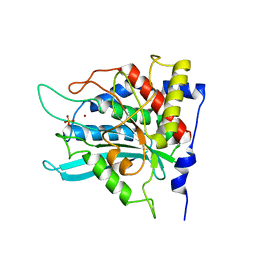 | | Crystal structure of the human glutaminyl cyclase mutant E201Q at 1.8 angstrom resolution | | Descriptor: | Glutaminyl-peptide cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Huang, K.F, Wang, Y.R, Chang, E.C, Chou, T.L, Wang, A.H. | | Deposit date: | 2007-12-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A conserved hydrogen-bond network in the catalytic centre of animal glutaminyl cyclases is critical for catalysis.
Biochem.J., 411, 2008
|
|
1EY3
 
 | |
5LO4
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPa-CH3 | | Authors: | Baker, E.G, Williams, C, Hudson, K.L, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
3F83
 
 | |
2JV6
 
 | | YF ED3 Protein NMR Structure | | Descriptor: | Envelope protein E | | Authors: | Volk, D.E, Gandham, S.H.A, May, F.J, Anderson, A, Barrett, A.D.T, Gorenstein, D.G. | | Deposit date: | 2007-09-12 | | Release date: | 2008-09-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Yellow Fever Virus Envelope Protein Domain III: A Convergence of Structure and Phylogenetics
To be Published
|
|
3NXX
 
 | | Preferential Selection of Isomer Binding from Chiral Mixtures: Alternate Binding Modes Observed for the E- and Z-isomers of a Series of 5-Substituted 2,4-Diaminofuro-2,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | Descriptor: | 5-[(1E)-2-(2-methoxyphenyl)-4-methylpent-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2K06
 
 | |
2M3J
 
 | |
1PD1
 
 | | Crystal structure of the COPII coat subunit, Sec24, complexed with a peptide containing the DxE cargo sorting signal of yeast Sys1 protein | | Descriptor: | DxE cargo sorting signal peptide of yeast Sys1 protein, Protein transport protein Sec24, ZINC ION | | Authors: | Mossessova, E, Bickford, L.C, Goldberg, J. | | Deposit date: | 2003-05-18 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | SNARE selectivity of the COPII coat.
Cell(Cambridge,Mass.), 114, 2003
|
|
1PD0
 
 | | Crystal structure of the COPII coat subunit, Sec24, complexed with a peptide from the SNARE protein Sed5 (yeast syntaxin-5) | | Descriptor: | COPII-binding peptide of the integral membrane protein SED5, Protein transport protein Sec24, ZINC ION | | Authors: | Mossessova, E, Bickford, L.C, Goldberg, J. | | Deposit date: | 2003-05-18 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | SNARE selectivity of the COPII coat.
Cell(Cambridge,Mass.), 114, 2003
|
|
3NZK
 
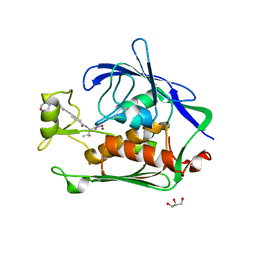 | | Structure of LpxC from Yersinia enterocolitica Complexed with CHIR090 Inhibitor | | Descriptor: | GLYCEROL, N-{(1S,2R)-2-hydroxy-1-[(hydroxyamino)carbonyl]propyl}-4-{[4-(morpholin-4-ylmethyl)phenyl]ethynyl}benzamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Cole, K.E, Christianson, D.W. | | Deposit date: | 2010-07-16 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Metal-Dependent Deacetylase LpxC from Yersinia enterocolitica Complexed with the Potent Inhibitor CHIR-090 .
Biochemistry, 50, 2010
|
|
6EK1
 
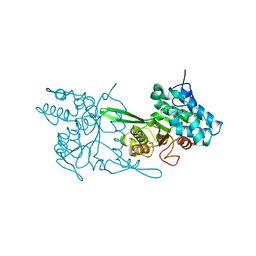 | | Crystal structure of Type IIP restriction endonuclease PfoI | | Descriptor: | 1,2-ETHANEDIOL, restriction endonuclease PfoI | | Authors: | Tamulaitiene, G, Manakova, E, Jovaisaite, V, Grazulis, S, Siksnys, V. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Unique mechanism of target recognition by PfoI restriction endonuclease of the CCGG-family.
Nucleic Acids Res., 47, 2019
|
|
5FEQ
 
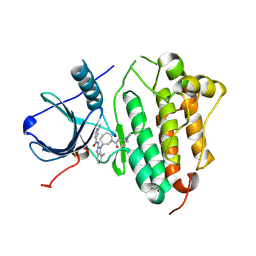 | | EGFR KINASE DOMAIN IN COMPLEX WITH A COVALENT AMINOBENZIMIDAZOLE | | Descriptor: | Epidermal growth factor receptor, ~{N}-[1-[(3~{R})-1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl]-7-methyl-benzimidazol-2-yl]-2-methyl-pyridine-4-carboxamide | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2015-12-17 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Discovery of (R,E)-N-(7-Chloro-1-(1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl)-1H-benzo[d]imidazol-2-yl)-2-methylisonicotinamide (EGF816), a Novel, Potent, and WT Sparing Covalent Inhibitor of Oncogenic (L858R, ex19del) and Resistant (T790M) EGFR Mutants for the Treatment of EGFR Mutant Non-Small-Cell Lung Cancers.
J.Med.Chem., 59, 2016
|
|
1DPS
 
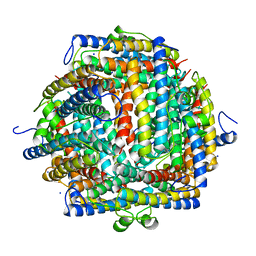 | | THE CRYSTAL STRUCTURE OF DPS, A FERRITIN HOMOLOG THAT BINDS AND PROTECTS DNA | | Descriptor: | DPS, SODIUM ION | | Authors: | Grant, R.A, Filman, D.J, Finkel, S.E, Kolter, R, Hogle, J.M. | | Deposit date: | 1998-02-23 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Dps, a ferritin homolog that binds and protects DNA.
Nat.Struct.Biol., 5, 1998
|
|
3RLU
 
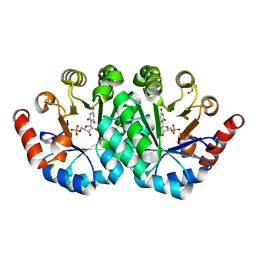 | | Crystal structure of the mutant K82A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-04-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
2MFI
 
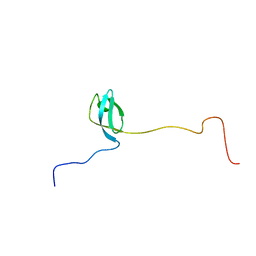 | | Domain 1 of E. coli ribosomal protein S1 | | Descriptor: | 30S ribosomal protein S1 | | Authors: | Giraud, P, Crechet, J, Bontems, F, Uzan, M, Sizun, C. | | Deposit date: | 2013-10-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Resonance assignment of the ribosome binding domain of E. coli ribosomal protein S1.
Biomol.Nmr Assign., 9, 2015
|
|
2MFL
 
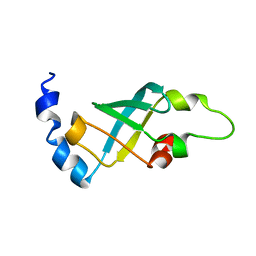 | | Domain 2 of E. coli ribosomal protein S1 | | Descriptor: | 30S ribosomal protein S1 | | Authors: | Giraud, P, Crechet, J, Bontems, F, Uzan, M, Sizun, C. | | Deposit date: | 2013-10-12 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Resonance assignment of the ribosome binding domain of E. coli ribosomal protein S1.
Biomol.Nmr Assign., 9, 2015
|
|
3F64
 
 | | F17a-G lectin domain with bound GlcNAc(beta1-O)paranitrophenyl ligand | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, F17a-G | | Authors: | Buts, L, De Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-05 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli.
Biology (Basel), 2, 2013
|
|
1ICV
 
 | | THE STRUCTURE OF ESCHERICHIA COLI NITROREDUCTASE COMPLEXED WITH NICOTINIC ACID | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Lovering, A.L, Hyde, E.I, Searle, P.F, White, S.A. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of Escherichia coli nitroreductase complexed with nicotinic acid: three crystal forms at 1.7 A, 1.8 A and 2.4 A resolution.
J.Mol.Biol., 309, 2001
|
|
5LO2
 
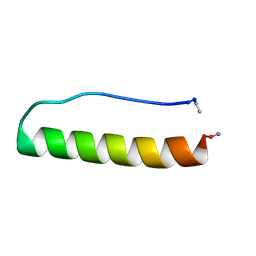 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPaTyr | | Authors: | Baker, E.G, Hudson, K.L, Williams, C, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
5LO3
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPaOMe | | Authors: | Baker, E.G, Hudson, K.L, Williams, C, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
3FNV
 
 | | Crystal Structure of Miner1: The Redox-active 2Fe-2S Protein Causative in Wolfram Syndrome 2 | | Descriptor: | CDGSH iron sulfur domain-containing protein 2, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Conlan, A.R, Axelrod, H.L, Cohen, A.E, Abresch, E.C, Yee, D, Zuris, J, Nechushtai, R, Jennings, P.A, Paddock, M.L. | | Deposit date: | 2008-12-26 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Miner1: The redox-active 2Fe-2S protein causative in Wolfram Syndrome 2.
J.Mol.Biol., 392, 2009
|
|
