3BQ0
 
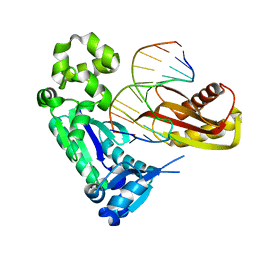 | | Pre-insertion binary complex of Dbh DNA polymerase | | Descriptor: | CALCIUM ION, DNA (5'-D(*DGP*DAP*DAP*DGP*DCP*DCP*DGP*DGP*DCP*DG)-3'), DNA (5'-D(*DT*DTP*DCP*DCP*DGP*DCP*DCP*DCP*DGP*DGP*DCP*DTP*DTP*DCP*DC)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2007-12-19 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh.
Mol.Cell, 29, 2008
|
|
3BQ2
 
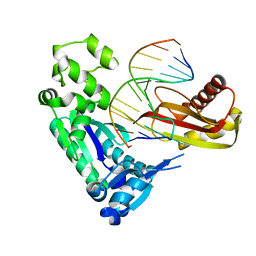 | | Post-insertion binary complex of Dbh DNA polymerase | | Descriptor: | CALCIUM ION, DNA (5'-D(*DGP*DAP*DAP*DGP*DCP*DCP*DGP*DGP*DCP*DGP*DG)-3'), DNA (5'-D(*DTP*DTP*DCP*DCP*DGP*DCP*DCP*DCP*DGP*DGP*DCP*DTP*DTP*DCP*DC)-3'), ... | | Authors: | Pata, J.D, Wilson, R.C. | | Deposit date: | 2007-12-19 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh.
Mol.Cell, 29, 2008
|
|
1WTP
 
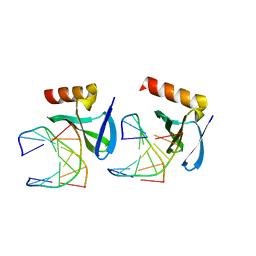 | | Hyperthermophile chromosomal protein SAC7D single mutant M29F in complex with DNA GCGA(UBr)CGC | | Descriptor: | 5'-D(*GP*CP*GP*AP*(BRU)P*CP*GP*C)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Chen, C.-Y, Ko, T.-P, Lin, T.-W, Chou, C.-C, Chen, C.-J, Wang, A.H.-J. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
NUCLEIC ACIDS RES., 33, 2005
|
|
1WTR
 
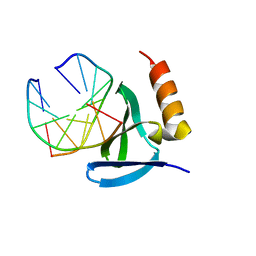 | | Hyperthermophile chromosomal protein SAC7D single mutant M29A in complex with DNA GCGATCGC | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Chen, C.-Y, Ko, T.-P, Lin, T.-W, Chou, C.-C, Chen, C.-J, Wang, A.H.-J. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
NUCLEIC ACIDS RES., 33, 2005
|
|
1WTW
 
 | | Hyperthermophile chromosomal protein SAC7D single mutant V26A in complex with DNA GCGATCGC | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Chen, C.-Y, Ko, T.-P, Lin, T.-W, Chou, C.-C, Chen, C.-J, Wang, A.H.-J. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
NUCLEIC ACIDS RES., 33, 2005
|
|
1WTQ
 
 | | Hyperthermophile chromosomal protein SAC7D single mutant M29F in complex with DNA GTAATTAC | | Descriptor: | 5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Chen, C.-Y, Ko, T.-P, Lin, T.-W, Chou, C.-C, Chen, C.-J, Wang, A.H.-J. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
NUCLEIC ACIDS RES., 33, 2005
|
|
1URJ
 
 | | Single stranded DNA-binding protein(ICP8) from Herpes simplex virus-1 | | Descriptor: | MAJOR DNA-BINDING PROTEIN, MERCURY (II) ION, ZINC ION | | Authors: | Panjikar, S, Mapelli, M, Tucker, P.A. | | Deposit date: | 2003-10-30 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the herpes simplex virus 1 ssDNA-binding protein suggests the structural basis for flexible, cooperative single-stranded DNA binding.
J. Biol. Chem., 280, 2005
|
|
7W1M
 
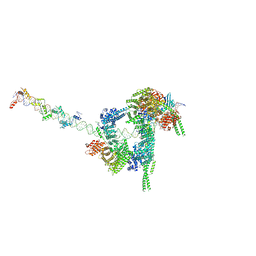 | | Cryo-EM structure of human cohesin-CTCF-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cohesin subunit SA-1, ... | | Authors: | Shi, Z.B, Bai, X.C, Yu, H. | | Deposit date: | 2021-11-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | CTCF and R-loops are boundaries of cohesin-mediated DNA looping.
Mol.Cell, 83, 2023
|
|
3JPP
 
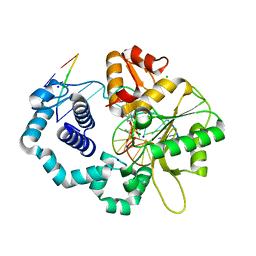 | | Ternary complex of DNA polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-MonoMethyl Methylene triphosphate | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy({(S)-hydroxy[(1R)-1-phosphonoethyl]phosphoryl}oxy)phosphoryl]guanosine, 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3', ... | | Authors: | Batra, V.K, Upton, J, Kashmerov, B, Beard, W.A, Wilson, S.H, Goodman, M.F, McKenna, C.E. | | Deposit date: | 2009-09-04 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Halogenated beta,gamma-Methylene- and Ethylidene-dGTP-DNA Ternary Complexes with DNA Polymerase beta: Structural Evidence for Stereospecific Binding of the Fluoromethylene Analogues.
J.Am.Chem.Soc., 132, 2010
|
|
6Q4V
 
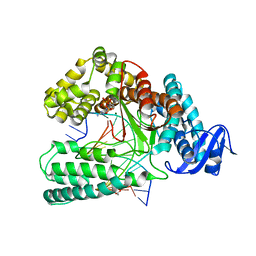 | | KlenTaq DNA polymerase in complex with dATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*CP*TP*GP*TP*GP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | The Structure of an Archaeal B-Family DNA Polymerase in Complex with a Chemically Modified Nucleotide.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
7P30
 
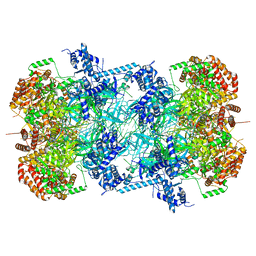 | | 3.0 A resolution structure of a DNA-loaded MCM double hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (53-MER), ... | | Authors: | Greiwe, J.F, Miller, T.C.R, Martino, F, Costa, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-02-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mechanism for the selective phosphorylation of DNA-loaded MCM double hexamers by the Dbf4-dependent kinase.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3EI2
 
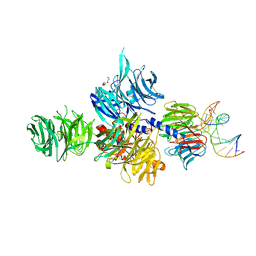 | | Structure of hsDDB1-drDDB2 bound to a 16 bp abasic site containing DNA-duplex | | Descriptor: | 5'-D(*DAP*DAP*DAP*DTP*DGP*DAP*DAP*DTP*(3DR)P*DAP*DAP*DGP*DCP*DAP*DGP*DG)-3', 5'-D(*DCP*DCP*DTP*DGP*DCP*DTP*DTP*DTP*DAP*DTP*DTP*DCP*DAP*DTP*DTP*DT)-3', DNA damage-binding protein 1, ... | | Authors: | Scrima, A, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
6Q4U
 
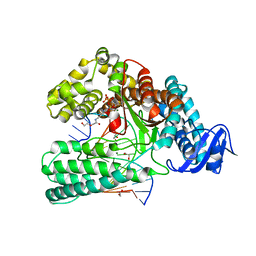 | | KlenTaq DNA pol in a closed ternary complex with 7-deaza-7-(2-(2-hydroxyethoxy)-N-(prop-2-yn-1-yl)acetamide)-2-dATP | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*CP*TP*GP*TP*GP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*CP*AP*(DOC))-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | The Structure of an Archaeal B-Family DNA Polymerase in Complex with a Chemically Modified Nucleotide.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
3EI1
 
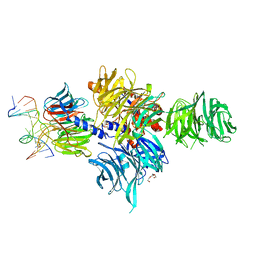 | |
4E5Z
 
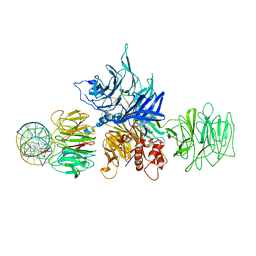 | |
8EBT
 
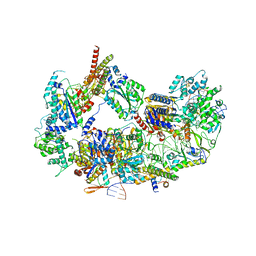 | |
6FBR
 
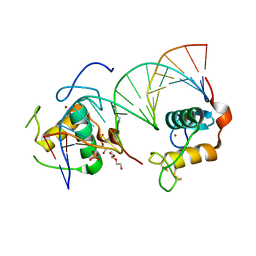 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human MEp DR1 Response Element, pH 4.2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*TP*GP*GP*GP*TP*CP*AP*AP*AP*GP*TP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*AP*CP*TP*TP*TP*GP*AP*CP*CP*CP*AP*G)-3'), ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2017-12-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modulation of RXR-DNA complex assembly by DNA context.
Mol. Cell. Endocrinol., 481, 2019
|
|
6FBQ
 
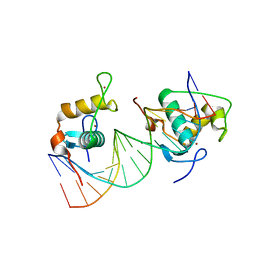 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human MEp DR1 Response Element, pH 7.0 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, DNA (5'-D(*CP*TP*GP*GP*GP*TP*CP*AP*AP*AP*GP*TP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*AP*CP*TP*TP*TP*GP*AP*CP*CP*CP*AP*G)-3'), ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2017-12-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Modulation of RXR-DNA complex assembly by DNA context.
Mol. Cell. Endocrinol., 481, 2019
|
|
4GCL
 
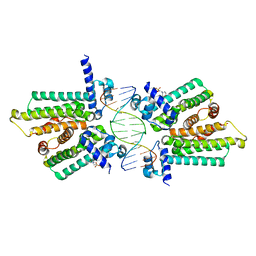 | | structure of no-dna factor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*GP*TP*GP*AP*GP*TP*AP*CP*TP*CP*AP*CP*T)-3'), Nucleoid occlusion factor SlmA | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-07-30 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8CIY
 
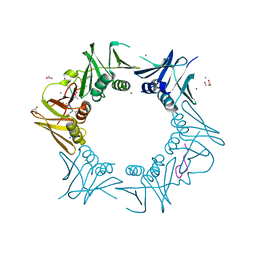 | | DNA-polymerase sliding clamp (DnaN) from Escherichia coli in complex with Cyclohexyl-Griselimycin. | | Descriptor: | ACETATE ION, Beta sliding clamp, CALCIUM ION, ... | | Authors: | Fu, C, Liu, Y, Walt, C, Bader, C, Rasheed, S, Lukat, P, Neuber, M, Blankenfeldt, W, Kalinina, O, Mueller, R. | | Deposit date: | 2023-02-11 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Elucidation of unusual biosynthesis and DnaN-targeting mode of action of potent anti-tuberculosis antibiotics Mycoplanecins.
Nat Commun, 15, 2024
|
|
8CIZ
 
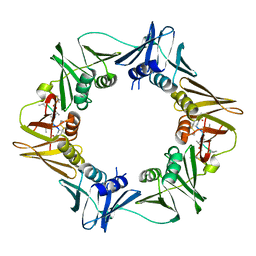 | | DNA-polymerase sliding clamp (DnaN) from Escherichia coli in complex with Mycoplanecin A. | | Descriptor: | Beta sliding clamp, Mycoplanecin A | | Authors: | Fu, C, Liu, Y, Walt, C, Bader, C, Rasheed, S, Lukat, P, Neuber, M, Blankenfeldt, W, Kalinina, O, Mueller, R. | | Deposit date: | 2023-02-11 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Elucidation of unusual biosynthesis and DnaN-targeting mode of action of potent anti-tuberculosis antibiotics Mycoplanecins.
Nat Commun, 15, 2024
|
|
8CIX
 
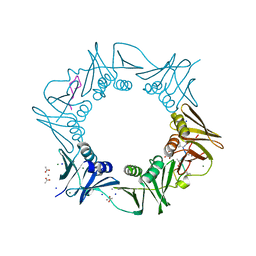 | | DNA-polymerase sliding clamp (DnaN) from Escherichia coli in complex with Griselimycin. | | Descriptor: | ACETATE ION, Beta sliding clamp, CALCIUM ION, ... | | Authors: | Fu, C, Liu, Y, Walt, C, Bader, C, Rasheed, S, Lukat, P, Neuber, M, Blankenfeldt, W, Kalinina, O, Mueller, R. | | Deposit date: | 2023-02-11 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Elucidation of unusual biosynthesis and DnaN-targeting mode of action of potent anti-tuberculosis antibiotics Mycoplanecins.
Nat Commun, 15, 2024
|
|
1AW4
 
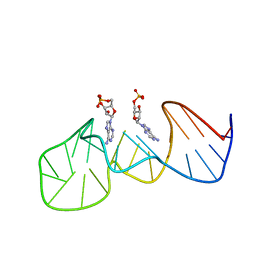 | |
8B9B
 
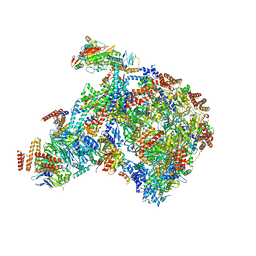 | |
8B9A
 
 | |
