5MWI
 
 | |
5MWN
 
 | | Structure of the EAEC T6SS component TssK N-terminal domain in complex with llama nanobodies nbK18 and nbK27 | | Descriptor: | PHOSPHATE ION, Type VI secretion protein, llama nanobody raised against TssK, ... | | Authors: | Cambillau, C, Nguyen, V.S, Spinelli, S, Legrand, P. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
5MXG
 
 | | Photorhabdus asymbiotica lectin (PHL) in complex with blood group H trisaccharide | | Descriptor: | Photorhabdus asymbiotica lectin PHL, SODIUM ION, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Jancarikova, G, Houser, J, Demo, G, Wimmerova, M. | | Deposit date: | 2017-01-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of novel bangle lectin from Photorhabdus asymbiotica with dual sugar-binding specificity and its effect on host immunity.
PLoS Pathog., 13, 2017
|
|
7MPE
 
 | |
2GPW
 
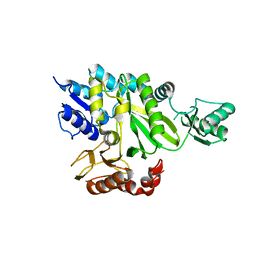 | | Crystal Structure of the Biotin Carboxylase Subunit, F363A Mutant, of Acetyl-CoA Carboxylase from Escherichia coli. | | Descriptor: | Biotin carboxylase | | Authors: | Shen, Y, Chou, C.Y, Chang, G.G, Tong, L. | | Deposit date: | 2006-04-18 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Is dimerization required for the catalytic activity of bacterial biotin carboxylase?
Mol.Cell, 22, 2006
|
|
2GLJ
 
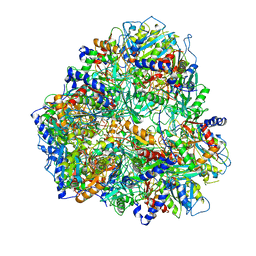 | |
7MT0
 
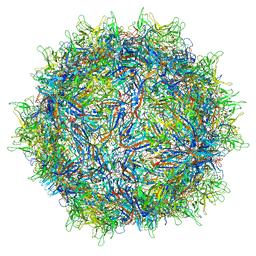 | | Structure of the adeno-associated virus 9 capsid at pH 7.4 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-12 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
2GQX
 
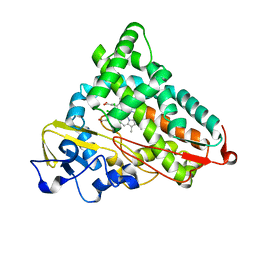 | | Crystal structure of cytochrome p450cam mutant (f87w/y96f/l244a/v247l/c334a) with pentachlorobenzene | | Descriptor: | 1,2,3,4,5-PENTACHLOROBENZENE, Cytochrome P450-cam, POTASSIUM ION, ... | | Authors: | Rao, Z, Wong, L.L, Xu, F, Bell, S.G. | | Deposit date: | 2006-04-22 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity correlations in pentachlorobenzene oxidation by engineered cytochrome P450cam
Protein Eng.Des.Sel., 20, 2007
|
|
2Z2T
 
 | |
2PVF
 
 | |
5MTU
 
 | | Maltodextrin binding protein MalE1 from L. casei BL23 bound to alpha-cyclodextrin | | Descriptor: | Cyclohexakis-(1-4)-(alpha-D-glucopyranose), MalE1 | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2017-01-10 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.999 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
2PVS
 
 | | Structure of human pancreatic lipase related protein 2 mutant N336Q | | Descriptor: | CALCIUM ION, Pancreatic lipase-related protein 2, SULFATE ION | | Authors: | Spinelli, S, Eydoux, C, Carriere, F, Cambillau, C. | | Deposit date: | 2007-05-10 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human pancreatic lipase-related protein 2 with the lid in an open conformation.
Biochemistry, 47, 2008
|
|
7MOA
 
 | | Cryo-EM structure of the c-MET II/HGF I complex bound with HGF II in a rigid conformation | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7M31
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Thymine and NADPH Anaerobically | | Descriptor: | Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Butrin, A, Beaupre, B, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Perturbing the Movement of Hydrogens to Delineate and Assign Events in the Reductive Activation and Turnover of Porcine Dihydropyrimidine Dehydrogenase.
Biochemistry, 60, 2021
|
|
7GHX
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-0e5afe9d-1 (Mpro-P0030) | | Descriptor: | (4R)-6-chloro-N-(1-methyl-2-oxo-2,3-dihydro-1H-imidazo[4,5-c]pyridin-7-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7MTZ
 
 | | Structure of the adeno-associated virus 9 capsid at pH pH 7.4 in complex with terminal galactose | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
2Z2B
 
 | |
5MV3
 
 | | ACC1 Fab fragment in complex with CII583-591 (CG10) | | Descriptor: | heavy chain of ACC1 Fab fragment, light chain of ACC1 Fab fragment, synthetic peptide containing the CII583-591 epitope of collagen type II,Collagen alpha-1(II) chain,synthetic peptide containing the CII583-591 epitope of collagen type II, | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-15 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
7MOB
 
 | | Cryo-EM structure of 2:2 c-MET/NK1 complex | | Descriptor: | Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
5MYA
 
 | | Homodimerization of Tie2 Fibronectin-like domains 1-3 in space group C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiopoietin-1 receptor, ... | | Authors: | Leppanen, V.-M, Saharinen, P, Alitalo, K. | | Deposit date: | 2017-01-26 | | Release date: | 2017-04-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of Tie2 activation and Tie2/Tie1 heterodimerization.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2Z3W
 
 | | Prolyl tripeptidyl aminopeptidase mutant E636A | | Descriptor: | Dipeptidyl aminopeptidase IV, GLYCEROL, SULFATE ION | | Authors: | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2007-06-07 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
5MYK
 
 | |
7MO7
 
 | | Cryo-EM structure of 2:2 c-MET/HGF holo-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
2Z4I
 
 | | Crystal structure of the Cpx pathway activator NlpE from Escherichia coli | | Descriptor: | Copper homeostasis protein cutF, HEXAETHYLENE GLYCOL, SULFATE ION | | Authors: | Hirano, Y, Hossain, M.M, Takeda, K, Tokuda, H, Miki, K. | | Deposit date: | 2007-06-18 | | Release date: | 2007-09-04 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies of the Cpx Pathway Activator NlpE on the Outer Membrane of Escherichia coli
Structure, 15, 2007
|
|
7M32
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671A Mutant Soaked with Uracil and NADPH Anaerobically | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ALANINE, Dihydropyrimidine dehydrogenase [NADP(+)], ... | | Authors: | Butrin, A, Beaupre, B, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Perturbing the Movement of Hydrogens to Delineate and Assign Events in the Reductive Activation and Turnover of Porcine Dihydropyrimidine Dehydrogenase.
Biochemistry, 60, 2021
|
|
