3SA8
 
 | | Crystal structure of wild-type HIV-1 protease in complex with KB83 | | Descriptor: | 3-hydroxy-N-[(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]benzamide, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
2BCE
 
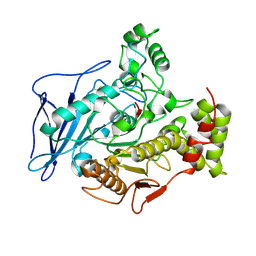 | | CHOLESTEROL ESTERASE FROM BOS TAURUS | | Descriptor: | CHOLESTEROL ESTERASE | | Authors: | Chen, J.C.-H, Miercke, L.J.W, Krucinski, J, Starr, J.R, Saenz, G, Wang, X, Spilburg, C.A, Lange, L.G, Ellsworth, J.L, Stroud, R.M. | | Deposit date: | 1998-01-28 | | Release date: | 1999-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of bovine pancreatic cholesterol esterase at 1.6 A: novel structural features involved in lipase activation.
Biochemistry, 37, 1998
|
|
2L6N
 
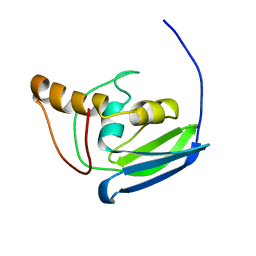 | | NMR solution structure of the protein YP_001092504.1 | | Descriptor: | uncharacterized protein YP_001092504.1 | | Authors: | Mohanty, B, Serrano, P, Geralt, M, Horst, R, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-11-23 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the protein YP_001092504.1
To be Published
|
|
3SA7
 
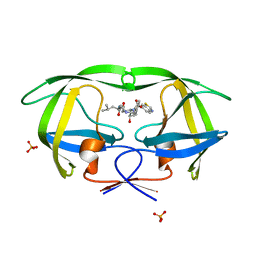 | | Crystal structure of wild-type HIV-1 protease in complex with AF55 | | Descriptor: | N~2~-acetyl-N-[(2S,3R)-4-{(1,3-benzothiazol-6-ylsulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]-L-leucinamide, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
3ODU
 
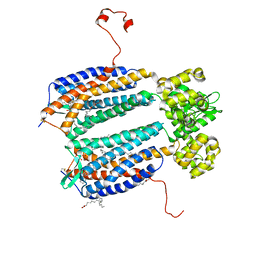 | | The 2.5 A structure of the CXCR4 chemokine receptor in complex with small molecule antagonist IT1t | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3OE6
 
 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in I222 spacegroup | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
1FXU
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM CALF SPLEEN IN COMPLEX WITH N(7)-ACYCLOGUANOSINE INHIBITOR AND A PHOSPHATE ION | | Descriptor: | 2-AMINO-7-[2-(2-HYDROXY-1-HYDROXYMETHYL-ETHYLAMINO)-ETHYL]-1,7-DIHYDRO-PURIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Luic, M, Bzowska, A, Shugar, D, Saenger, W, Koellner, G. | | Deposit date: | 2000-09-27 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase: structure of its ternary complex with an N(7)-acycloguanosine inhibitor and a phosphate anion.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2O0E
 
 | |
3T3G
 
 | | Glycogen Phosphorylase b in complex with GlcBrU | | Descriptor: | 5-bromo-1-(beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
2DFN
 
 | | Structure of shikimate kinase from Mycobacterium tuberculosis complexed with ADP and shikimate at 1.9 angstrons of resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Dias, M.V, Faim, L.M, Vasconcelos, I.B, de Oliveira, J.S, Basso, L.A, Santos, D.S, de Azevedo, W.F. | | Deposit date: | 2006-03-03 | | Release date: | 2006-09-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Effects of the magnesium and chloride ions and shikimate on the structure of shikimate kinase from Mycobacterium tuberculosis
ACTA CRYSTALLOGR.,SECT.F, 63, 2007
|
|
3DMK
 
 | | Crystal structure of Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, N-terminal eight Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, ... | | Authors: | Sawaya, M.R, Wojtowicz, W.M, Eisenberg, D, Zipursky, S.L. | | Deposit date: | 2008-07-01 | | Release date: | 2008-10-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.19 Å) | | Cite: | A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms.
Cell(Cambridge,Mass.), 134, 2008
|
|
2DIT
 
 | | Solution structure of the RRM_1 domain of HIV TAT specific factor 1 variant | | Descriptor: | HIV TAT specific factor 1 variant | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM_1 domain of HIV TAT specific factor 1 variant
To be Published
|
|
4CIW
 
 | | Crystal structure of Mycobacterium tuberculosis type 2 dehydroquinase in complex with (1R,4R,5R)-1,4,5-trihydroxy-3-(2-hydroxy)ethylcyclohex-2-ene-1-carboxylic acid | | Descriptor: | (1R,4R,5R)-1,4,5-trihydroxy-3-(2-hydroxy)ethylcyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, SODIUM ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Lamb, H, Hawkins, A.R, Blanco, B, Sedes, A, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2013-12-17 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the water-binding pocket of the type II dehydroquinase enzyme in the structure-based design of inhibitors.
J. Med. Chem., 57, 2014
|
|
5EJ1
 
 | | Pre-translocation state of bacterial cellulose synthase | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ... | | Authors: | Moragn, J.L.W, Zimmer, J. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Observing cellulose biosynthesis and membrane translocation in crystallo.
Nature, 531, 2016
|
|
2O0D
 
 | |
2YJX
 
 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | 1-(3H-imidazo[4,5-c]pyridin-2-yl)-3,4-dihydropyrido[2,1-a]isoindol-6(2H)-one, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-24 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
1HI6
 
 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE | | Descriptor: | IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN), IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN), PEPTIDE 5 | | Authors: | Hahn, M, Wessner, H, Schneider-Mergener, J, Hohne, W. | | Deposit date: | 2001-01-02 | | Release date: | 2001-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic Analysis of Anti-P24 (HIV-1) Monoclonal Antibody Cross-Reactivity and Polyspecificity
Cell(Cambridge,Mass.), 91, 1997
|
|
3SAC
 
 | | Crystal structure of wild-type HIV-1 protease in complex with AF80 | | Descriptor: | 3-hydroxy-N-{(2S,3R)-3-hydroxy-4-[(2-methylpropyl){[5-(1,2-oxazol-5-yl)thiophen-2-yl]sulfonyl}amino]-1-phenylbutan-2-yl}benzamide, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
3SAN
 
 | | Crystal structure of influenza A virus neuraminidase N5 complexed with Zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, M.Y, Qi, J.X, Liu, Y, Vavricka, C.J, Wu, Y, Li, Q, Gao, G.F. | | Deposit date: | 2011-06-03 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Influenza a virus n5 neuraminidase has an extended 150-cavity
J.Virol., 85, 2011
|
|
4GPO
 
 | |
3SA9
 
 | | Crystal structure of Wild-type HIV-1 protease in complex With AF68 | | Descriptor: | ACETATE ION, N~2~-acetyl-N-[(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]-L-isoleucinamide, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
4EYB
 
 | | Crystal structure of NDM-1 bound to hydrolyzed oxacillin | | Descriptor: | (2R,4S)-2-[(R)-carboxy{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carbo xylic acid, Beta-lactamase NDM-1, ZINC ION | | Authors: | Strynadka, N.C.J, King, D.T. | | Deposit date: | 2012-05-01 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | New Delhi Metallo-Beta-Lactamase: Structural Insights into Beta-Lactam Recognition and Inhibition
J.Am.Chem.Soc., 134, 2012
|
|
1HP4
 
 | |
2C54
 
 | | gdp-mannose-3', 5' -epimerase (arabidopsis thaliana),k178r, with gdp-beta-l-gulose and gdp-4-keto-beta-l-gulose bound in active site. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, GDP-MANNOSE-3', ... | | Authors: | Major, L.L, Wolucka, B.A, Naismith, J.H. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of GDP-mannose-3',5'-epimerase: an enzyme which performs three chemical reactions at the same active site.
J. Am. Chem. Soc., 127, 2005
|
|
2J08
 
 | | Thermus DNA photolyase with 8-Iod-riboflavin antenna chromophore | | Descriptor: | 1-DEOXY-1-(8-IODO-7-METHYL-2,4-DIOXO-3,4-DIHYDROBENZO[G]PTERIDIN-10(2H)-YL)-D-RIBITOL, CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
