3VUP
 
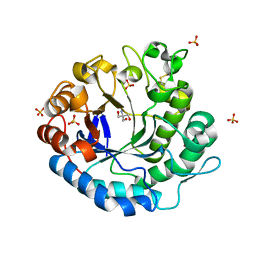 | | Beta-1,4-mannanase from the common sea hare Aplysia kurodai | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-1,4-mannanase, SULFATE ION | | Authors: | Mizutani, K, Tsuchiya, S, Toyoda, M, Nanbu, Y, Tominaga, K, Yuasa, K, Takahashi, N, Tsuji, A, Mikami, B. | | Deposit date: | 2012-07-04 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure of beta-1,4-mannanase from the common sea hare Aplysia kurodai at 1.05 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5UR4
 
 | |
4CQC
 
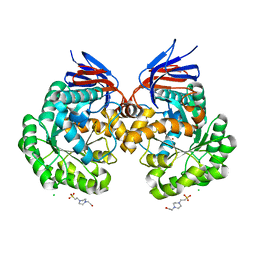 | | The reaction mechanism of the N-isopropylammelide isopropylaminohydrolase AtzC: insights from structural and mutagenesis studies | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, N-ISOPROPYLAMMELIDE ISOPROPYL AMIDOHYDROLASE, ... | | Authors: | Balotra, S, Newman, J, French, N.G, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-13 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
6BKZ
 
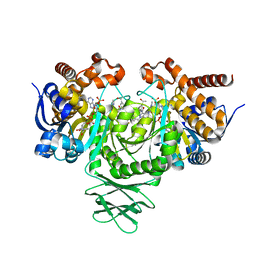 | | Novel Modes of Inhibition of Wild-Type IDH1: Non-equivalent Allosteric Inhibition with Cmpd3 | | Descriptor: | (7R)-1-[(4-fluorophenyl)methyl]-N-{3-[(1R)-1-hydroxyethyl]phenyl}-7-methyl-5-(1H-pyrrole-2-carbonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridine-3-carboxamide, Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2017-11-09 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315.
J. Med. Chem., 61, 2018
|
|
4QIJ
 
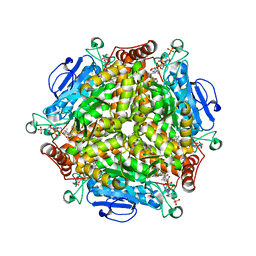 | | Crystal structure of MenB from Mycobacteria tuberculosis in complex with 1-HNA-CoA | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, 1-hydroxy-2-naphthoyl-CoA | | Authors: | Song, H.G, Sung, H.P, Tse, Y.S, Guo, Z.H. | | Deposit date: | 2014-05-31 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-dependent active-site closure revealed in the crystal structure of Mycobacterium tuberculosis MenB complexed with product analogues
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7KE5
 
 | | Heavy chain ferritin with N-terminal EBNA1 epitope | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Epstein-Barr nuclear antigen 1,Ferritin heavy chain, FE (III) ION | | Authors: | Pederick, J.P, Bruning, J.B. | | Deposit date: | 2020-10-10 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Immunogenicity study of engineered ferritins with C- and N-terminus insertion of Epstein-Barr nuclear antigen 1 epitope.
Vaccine, 39, 2021
|
|
7KE3
 
 | | Heavy chain ferritin with C-terminal EBNA1 epitope | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, Ferritin heavy chain,Epstein-Barr nuclear antigen 1 | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2020-10-10 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Immunogenicity study of engineered ferritins with C- and N-terminus insertion of Epstein-Barr nuclear antigen 1 epitope.
Vaccine, 39, 2021
|
|
6C4Q
 
 | | 1.16 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1-100) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Polyketide synthase Pks13 | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-12 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 1.16 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1-100) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis.
To Be Published
|
|
2CN0
 
 | | Complex of Recombinant Human Thrombin with a Designed Inhibitor | | Descriptor: | 4-(1R,3AS,4R,8AS,8BR)-[1-DIFLUOROMETHYL-2-(4-FLUOROBENZYL)-3-OXODECAHYDROPYRROLO[3,4-A]PYRROLIZIN-4-YL]BENZAMIDINE, CALCIUM ION, HIRUDIN IIA, ... | | Authors: | Hoffmann-Roder, A, Schweizer, E, Egger, J, Seiler, P, Obst-Sander, U, Wagner, B, Kansy, M, Banner, D.W, Diederich, F. | | Deposit date: | 2006-05-17 | | Release date: | 2006-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mapping the Fluorophilicity of a Hydrophobic Pocket: Synthesis and Biological Evaluation of Tricyclic Thrombin Inhibitors Directing Fluorinated Alkyl Groups Into the P Pocket
Chemmedchem, 1, 2006
|
|
6D4V
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 22 (VCC061422) | | Descriptor: | 2-cyclohexyl-1-{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}ethan-1-one, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
6D4U
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 27 (VCC663664) | | Descriptor: | 2-(2,4-dimethoxyphenyl)-1-{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}ethan-1-one, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
6D67
 
 | | Crystal structure of the human dual specificity phosphatase 1 catalytic domain (C258S) as a maltose binding protein fusion (maltose bound form) in complex with the designed AR protein mbp3_16 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Designed AR protein mbp3_16, ... | | Authors: | Gumpena, R, Lountos, G.T, Waugh, D.S. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | MBP-binding DARPins facilitate the crystallization of an MBP fusion protein.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5TQ1
 
 | |
4CMI
 
 | |
6CQ8
 
 | | K2P2.1(TREK-1):ML335 complex | | Descriptor: | CADMIUM ION, HEXADECANE, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2018-03-14 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | K2P2.1 (TREK-1)-activator complexes reveal a cryptic selectivity filter binding site.
Nature, 547, 2017
|
|
3G5E
 
 | | Human aldose reductase complexed with IDD 740 inhibitor | | Descriptor: | 2-(3-((4,5,7-trifluorobenzo[d]thiazol-2-yl)methyl)-1H-pyrrolo[2,3-b]pyridin-1-yl)acetic acid, Aldose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Podjarny, A.D, Van Zandt, M.C. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of [3-(4,5,7-trifluoro-benzothiazol-2-ylmethyl)-pyrrolo[2,3-b]pyridin-1-yl]acetic acids as highly potent and selective inhibitors of aldose reductase for treatment of chronic diabetic complications.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3M07
 
 | | 1.4 Angstrom Resolution Crystal Structure of Putative alpha Amylase from Salmonella typhimurium. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-02 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 1.4 Angstrom Resolution Crystal Structure of Putative alpha Amylase from Salmonella typhimurium.
TO BE PUBLISHED
|
|
6D4W
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 35 (VCC620637) | | Descriptor: | 2-(4-fluorophenyl)-1-{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}ethan-1-one, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
4CM6
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | (E)-2-amino-4-oxo-6-styryl-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CMK
 
 | |
5FDR
 
 | | Mcl-1 complexed with small molecule inhibitor | | Descriptor: | 5-[[6-chloranyl-3-[3-(4-chloranyl-3,5-dimethyl-phenoxy)propyl]-7-(3,5-dimethyl-1~{H}-pyrazol-4-yl)-1~{H}-indol-2-yl]carbonylsulfamoyl]furan-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2015-12-16 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 2-Indole-acylsulfonamide Myeloid Cell Leukemia 1 (Mcl-1) Inhibitors Using Fragment-Based Methods.
J.Med.Chem., 59, 2016
|
|
5OMY
 
 | | HIGH-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-02 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
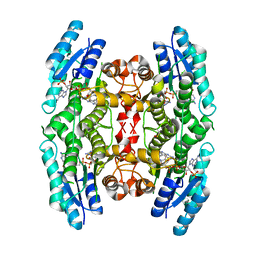
4CLR
 
 | |
4CMC
 
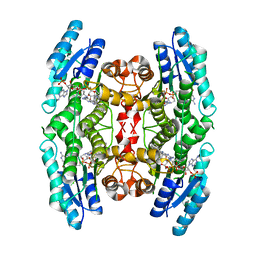 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | N4-cyclohexyl-5,6-diphenyl-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 1 | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4FFY
 
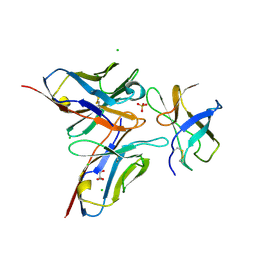 | | Crystal structure of DENV1-E111 single chain variable fragment bound to DENV-1 DIII, strain 16007. | | Descriptor: | CHLORIDE ION, DENV1-E111 single chain variable fragment (heavy chain), DENV1-E111 single chain variable fragment (light chain), ... | | Authors: | Austin, S.K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Differential Neutralization of DENV-1 Genotypes by an Antibody that Recognizes a Cryptic Epitope.
Plos Pathog., 8, 2012
|
|
