1C5G
 
 | | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Goldsmith, E.J. | | Deposit date: | 1999-12-07 | | Release date: | 1999-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering of plasminogen activator inhibitor-1 to reduce the rate of latency transition.
Nat.Struct.Biol., 2, 1995
|
|
2OGR
 
 | |
4DXA
 
 | | Co-crystal structure of Rap1 in complex with KRIT1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Krev interaction trapped protein 1, MAGNESIUM ION, ... | | Authors: | Li, X, Zhang, R, Boggon, T.J. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Small G Protein Effector Interaction of Ras-related Protein 1 (Rap1) and Adaptor Protein Krev Interaction Trapped 1 (KRIT1).
J.Biol.Chem., 287, 2012
|
|
1W1H
 
 | | Crystal Structure of the PDK1 Pleckstrin Homology (PH) domain | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, GLYCEROL, SULFATE ION | | Authors: | Komander, D, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2004-06-21 | | Release date: | 2004-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Insights Into the Regulation of Pdk1 by Phosphoinositides and Inositol Phosphates
Embo J., 23, 2004
|
|
2XT0
 
 | | Dehalogenase DPpA from Plesiocystis pacifica SIR-I | | Descriptor: | HALOALKANE DEHALOGENASE, SULFATE ION | | Authors: | Bogdanovic, X, Palm, G.J, Hinrichs, W. | | Deposit date: | 2010-10-02 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cloning, Functional Expression, Biochemical Characterization, and Structural Analysis of a Haloalkane Dehalogenase from Plesiocystis Pacifica Sir-1.
Appl.Microbiol.Biotechnol., 91, 2011
|
|
5T20
 
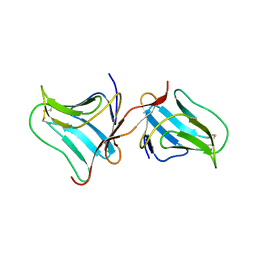 | | Crystal Structure of Tarin Lectin bound to Trimannose | | Descriptor: | 1,2-ETHANEDIOL, Lectin, alpha-D-mannopyranose, ... | | Authors: | Pereira, P.R, Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2016-08-22 | | Release date: | 2016-09-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | High-resolution crystal structures of Colocasia esculenta tarin lectin.
Glycobiology, 27, 2017
|
|
4J53
 
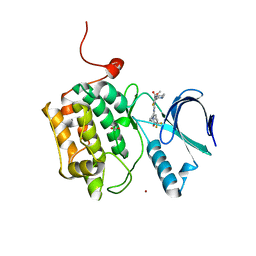 | | Crystal structure of PLK1 in complex with TAK-960 | | Descriptor: | 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-2-fluoro-5-methoxy-N-(1-methylpiperidin-4-yl)benzamide, ACETATE ION, Serine/threonine-protein kinase PLK1, ... | | Authors: | Hosfield, D.J, Skene, R.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of TAK-960: An orally available small molecule inhibitor of polo-like kinase 1 (PLK1).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HAW
 
 | |
2NS6
 
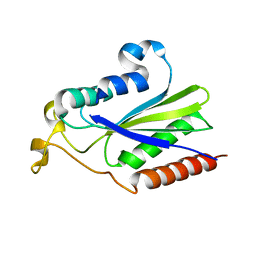 | | Crystal Structure of the Minimal Relaxase Domain of MobA from Plasmid R1162 | | Descriptor: | MANGANESE (II) ION, Mobilization protein A | | Authors: | Monzingo, A.F, Ozburn, A, Xia, S, Meyer, R.J, Robertus, J.D. | | Deposit date: | 2006-11-03 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the Minimal Relaxase Domain of MobA at 2.1 A Resolution.
J.Mol.Biol., 366, 2007
|
|
2Y62
 
 | | Crystal structure of Leishmanial E65Q-TIM complexed with R-Glycidol phosphate | | Descriptor: | GLYCEROL, SN-GLYCEROL-1-PHOSPHATE, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Venkatesan, R, Alahuhta, M, Pihko, P.M, Wierenga, R.K. | | Deposit date: | 2011-01-19 | | Release date: | 2011-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High Resolution Crystal Structures of Triosephosphate Isomerase Complexed with its Suicide Inhibitors: The Conformational Flexibility of the Catalytic Glutamate in its Closed, Liganded Active Site.
Protein Sci., 20, 2011
|
|
2N8N
 
 | | Solution structure of translation initiation factor | | Descriptor: | Translation initiation factor IF-1 | | Authors: | Kim, D, Lee, K, Lee, B. | | Deposit date: | 2015-10-21 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics study of translation initiation factor 1 from Staphylococcus aureus suggests its RNA binding mode
Biochim.Biophys.Acta, 1865, 2017
|
|
3NG4
 
 | | Ternary complex of peptidoglycan recognition protein (PGRP-S) with Maltose and N-Acetylglucosamine at 1.7 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Multiligand specificity of pathogen-associated molecular pattern-binding site in peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
4DRP
 
 | | Evaluation of Synthetic FK506 Analogs as Ligands for the FK506-Binding Proteins 51 and 52: Complex of FKBP51 with 2-(3-((R)-3-(3,4-dimethoxyphenyl)-1-((S)-1-(2-((1R,2S)-2-ethyl-1-hydroxy-cyclohexyl)-2-oxoacetyl)piperidine-2-carbonyloxy)propyl)phenoxy)acetic acid from cocrystallization | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1R,2S)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
4B0O
 
 | | Crystal structure of soman-aged human butyrylcholinesterase in complex with benzyl pyridinium-4-methyltrichloroacetimidate | | Descriptor: | 1-BENZYL-4-({[(1E)-2,2,2-TRICHLOROETHANIMIDOYL]OXY}METHYL)PYRIDINIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wandhammer, M, de Koning, M, van Grol, M, Noort, D, Goeldner, M, Nachon, F. | | Deposit date: | 2012-07-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Step Toward the Reactivation of Aged Cholinesterases -Crystal Structure of Ligands Binding to Aged Human Butyrylcholinesterase
Chem.Biol.Interact, 203, 2013
|
|
4H87
 
 | |
2J4O
 
 | | Structure of TAB1 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 7-INTERACTING PROTEIN 1 | | Authors: | van Aalten, D. | | Deposit date: | 2006-09-01 | | Release date: | 2006-09-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tak1-Binding Protein 1 is a Pseudophosphatase.
Biochem.J., 399, 2006
|
|
3NNC
 
 | | Crystal Structure of CUGBP1 RRM1/2-RNA Complex | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*UP*GP*UP*GP*UP*GP*UP*UP*GP*UP*GP*UP*G)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2005 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
4HB4
 
 | |
2IMD
 
 | | Structure of SeMet 2-hydroxychromene-2-carboxylate isomerase (HCCA isomerase) | | Descriptor: | (2S)-2-HYDROXY-2H-CHROMENE-2-CARBOXYLIC ACID, (3E)-4-(2-HYDROXYPHENYL)-2-OXOBUT-3-ENOIC ACID, 2-hydroxychromene-2-carboxylate isomerase, ... | | Authors: | Thompson, L.C, Ladner, J.E, Codreanu, S.G, Harp, J, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2006-10-04 | | Release date: | 2007-06-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 2-Hydroxychromene-2-carboxylic Acid Isomerase: A Kappa Class Glutathione Transferase from Pseudomonas putida
Biochemistry, 46, 2007
|
|
4D71
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 2 blood group A-tetrasaccharide (E558A X02 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, SULFATE ION, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-19 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
3N3X
 
 | | Crystal Structure of the complex formed between type I ribosome inactivating protein and hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GUANINE, Ribosome inactivating protein, ... | | Authors: | Kushwaha, G.S, Vikram, G, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the complex formed between type I ribosome inactivating protein and hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 1.7 A resolution
To be Published
|
|
3N49
 
 | | Human FPPS COMPLEX WITH NOV_292 | | Descriptor: | FARNESYL PYROPHOSPHATE SYNTHASE, PHOSPHATE ION, naphtho[2,1-b]thiophen-1-ylacetic acid | | Authors: | Rondeau, J.-M. | | Deposit date: | 2010-05-21 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery.
Nat.Chem.Biol., 6, 2010
|
|
2C4M
 
 | |
5ANN
 
 | | Structure of fructofuranosidase from Xanthophyllomyces dendrorhous | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-FRUCTOFURANOSIDASE, ... | | Authors: | Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2015-09-07 | | Release date: | 2016-02-10 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Analysis of Beta-Fructofuranosidase from Xanthophyllomyces Dendrorhous Reveals Unique Features and the Crucial Role of N-Glycosylation in Oligomerization and Activity
J.Biol.Chem., 291, 2016
|
|
4D85
 
 | | Crystal Structure of Human Beta Secretase in Complex with NVP-BVI151 | | Descriptor: | (3R,4S,5S)-3-[(3-tert-butylbenzyl)amino]-5-[(4,4,7'-trifluoro-1',2'-dihydrospiro[cyclohexane-1,3'-indol]-5'-yl)methyl]tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1, IODIDE ION | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2012-01-10 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of cyclic sulfone hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides
J.Med.Chem., 55, 2012
|
|
