6TE5
 
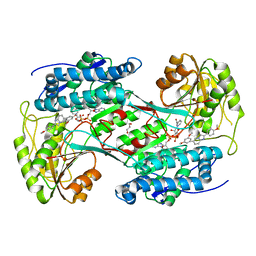 | | Crystal structure of human Aldehyde dehydrogenase 1A3 in complex with LQ43 inhibitor compound | | Descriptor: | 6-(3,5-dimethoxyphenyl)-2-(4-methoxyphenyl)imidazo[1,2-a]pyridine, Aldehyde dehydrogenase family 1 member A3, GLYCEROL, ... | | Authors: | Gelardi, E.L.M, Garavaglia, S. | | Deposit date: | 2019-11-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Imidazo[1,2- a ]pyridine Derivatives as Aldehyde Dehydrogenase Inhibitors: Novel Chemotypes to Target Glioblastoma Stem Cells.
J.Med.Chem., 63, 2020
|
|
6Y6F
 
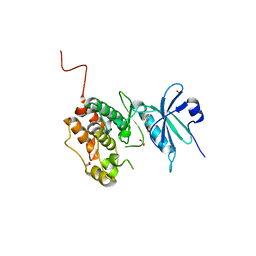 | | Crystal structure of STK17B (DRAK2) in complex with PKIS43 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(4-methylsulfanylphenyl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Drewry, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
7TB1
 
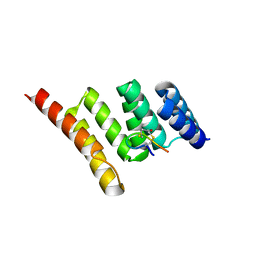 | | Crystal structure of STUB1 with a macrocyclic peptide | | Descriptor: | 1,3-bis(sulfanyl)propan-2-one, ALA-CYS-SER-SER-ILE-TRP-CYS-PRO-ASP-GLY, E3 ubiquitin-protein ligase CHIP | | Authors: | Bahmanjah, S, Klein, D.J. | | Deposit date: | 2021-12-21 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Discovery and Structure-Based Design of Macrocyclic Peptides Targeting STUB1.
J.Med.Chem., 2022
|
|
7KNN
 
 | |
6YD1
 
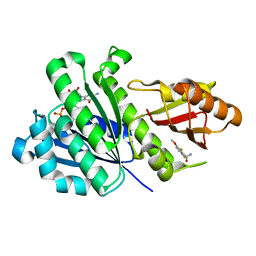 | | SaFtsZ-DFMBA | | Descriptor: | 2,6-difluoro-3-methoxybenzamide, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
6YD6
 
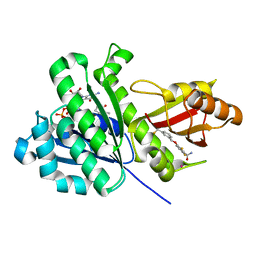 | | SaFtsZ-UCM152 (comp.20) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylpyrrolidin-2-one, 2,6-bis(fluoranyl)-3-[[3-(trifluoromethyl)phenyl]methoxy]benzamide, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
6YD5
 
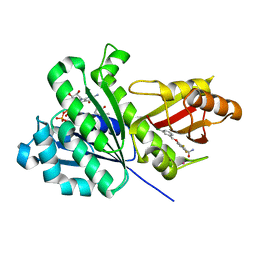 | | SaFtsZ-UCM151 (comp. 18) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylpyrrolidin-2-one, 3-[(3-chlorophenyl)methoxy]-2,6-bis(fluoranyl)benzamide, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
7KRY
 
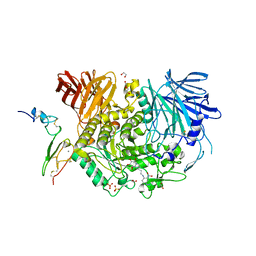 | | Co-crystal structure of alpha glucosidase with compound 11 | | Descriptor: | (1S,2S,3R,4S,5S)-5-({6-[(4-azido-2-nitrophenyl)amino]hexyl}amino)-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
2UU8
 
 | | X-ray structure of Ni, Ca concanavalin A at Ultra-high resolution (0. 94A) | | Descriptor: | CALCIUM ION, CONCANAVALIN, NICKEL (II) ION | | Authors: | Ahmed, H.U, Blakeley, M.P, Cianci, M, Cruickshank, D.W.J, Hubbard, J.A, Helliwell, J.R. | | Deposit date: | 2007-03-01 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | The Determination of Protonation States in Proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6HB7
 
 | | Crystal structure of E. coli tyrRS in complex with 5'-O-(N-L-tyrosyl)sulfamoyl-N3-methyluridine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Tyrosine--tRNA ligase, ... | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-08-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative analysis of pyrimidine substituted aminoacyl-sulfamoyl nucleosides as potential inhibitors targeting class I aminoacyl-tRNA synthetases.
Eur.J.Med.Chem., 173, 2019
|
|
6YTD
 
 | | CLK1 V324A mutant bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YTI
 
 | | CLK1 bound with ETH1610 (Cpd 17) | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase CLK1, methyl 9-[(2-fluoranyl-4-methoxy-phenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YTA
 
 | | CLK1 bound with imidazopyridazine (Cpd 1) | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-{6-[(CYCLOPROPYLMETHYL)AMINO]IMIDAZO[1,2-B]PYRIDAZIN-3-YL}PHENYL)ETHANONE, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YTG
 
 | | CLK1 bound with beta-carboline KH-CARB13 (Cpd 3) | | Descriptor: | (4~{S})-7,8-bis(chloranyl)-9-methyl-1-oxidanylidene-spiro[2,4-dihydropyrido[3,4-b]indole-3,4'-piperidine]-4-carbonitrile, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Huber, K, Bracher, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YTY
 
 | | CLK3 A319V mutant bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YTE
 
 | | CLK1 bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6ZB0
 
 | |
6ZB1
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH GSK620 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, ~{N}5-cyclopropyl-~{N}3-methyl-2-oxidanylidene-1-(phenylmethyl)pyridine-3,5-dicarboxamide | | Authors: | Chung, C. | | Deposit date: | 2020-06-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Optimization of a Novel, Weak Bromo and Extra Terminal Domain (BET) Bromodomain Fragment Ligand to a Potent and Selective Second Bromodomain (BD2) Inhibitor.
J.Med.Chem., 63, 2020
|
|
1L7G
 
 | | Crystal structure of E119G mutant influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
6ZJF
 
 | | Crystal structure of STK17B (DRAK2) in complex with AP-229 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(4-cyclopropylphenyl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Picado, A, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
7MLT
 
 | | Crystal structure of ricin A chain in complex with 5-(2-ethylphenyl)thiophene-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-ethylphenyl)thiophene-2-carboxylic acid, CHLORIDE ION, ... | | Authors: | Harijan, R.K, Li, X.P, Cao, B, Augeri, D, Bonanno, J.B, Almo, S.C, Tumer, N.E, Schramm, V.L. | | Deposit date: | 2021-04-29 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Structural Characterization of Ricin Inhibitors Targeting Ribosome Binding Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
7MSO
 
 | |
6YYN
 
 | | Structure of Cathepsin S in complex with Compound 14 | | Descriptor: | CITRATE ANION, Cathepsin S, SULFATE ION, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YYR
 
 | | Structure of Cathepsin S in complex with Compound 20b | | Descriptor: | (2~{R})-~{N}-(2-azanylideneethyl)-2-[2-(3-methyl-1,2-oxazol-5-yl)ethanoylamino]-3-(4-pyridin-2-ylpiperazin-1-yl)sulfonyl-propanamide, 1,2-ETHANEDIOL, CITRATE ANION, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YYP
 
 | | Structure of Cathepsin S in complex with Compound 2 | | Descriptor: | 1-(furan-2-ylmethyl)-5-(trifluoromethyl)benzimidazol-2-amine, ACETATE ION, Cathepsin S, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
