1WAK
 
 | | X-ray structure of SRPK1 | | Descriptor: | 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE SPRK1 | | Authors: | Ngo, J.C, Gullingsrud, J, Chakrabarti, S, Nolen, B, Aubol, B.E, Fu, X.D, Adams, J.A, Mccammon, J.A, Ghosh, G. | | Deposit date: | 2004-10-26 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Sr Protein Kinase 1 is Resilient to Inactivation.
Structure, 15, 2007
|
|
1WA6
 
 | | The structure of ACC oxidase | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE 1, FE (II) ION, PHOSPHATE ION, ... | | Authors: | Zhang, Z, Ren, J.-S, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2004-10-25 | | Release date: | 2005-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure and Mechanistic Implications of 1-Aminocyclopropane-1-Carboxylic Acid Oxidase (the Ethyling Forming Enzyme)
Chem.Biol., 11, 2004
|
|
1W4T
 
 | | X-ray crystallographic structure of Pseudomonas aeruginosa arylamine N-acetyltransferase | | Descriptor: | Arylamine N-acetyltransferase, SULFATE ION | | Authors: | Westwood, I.M, Holton, S.J, Rodrigues-Lima, F, Dupret, J.-M, Bhakta, S, Noble, M.E, Sim, E. | | Deposit date: | 2004-07-29 | | Release date: | 2005-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Expression, purification, characterization and structure of Pseudomonas aeruginosa arylamine N-acetyltransferase.
Biochem. J., 385, 2005
|
|
1W5L
 
 | | An anti-parallel to parallel switch. | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Leman, L.J, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2004-08-07 | | Release date: | 2004-09-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Coiled Coils at the Edge of Configurational Heterogeneity. Structural Analyses of Parallel and Antiparallel Homotetrameric Coiled Coils Reveal Configurational Sensitivity to a Single Solvent-Exposed Amino Acid Substitution.
Biochemistry, 45, 2006
|
|
5WBB
 
 | | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins - S112A | | Descriptor: | COPPER (II) ION, GLYCEROL, SULFATE ION, ... | | Authors: | Mann, S.I, Heinisch, T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
1W5V
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DI-3-FLUORO-BENZYL-GLUCARYL]-DI-[1-AMINO-INDAN-2-OL] | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
1W5O
 
 | | Stepwise introduction of zinc binding site into porphobilinogen synthase of Pseudomonas aeruginosa (mutations A129C, D131C and D139C) | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Frere, F, Reents, H, Schubert, W.-D, Heinz, D.W, Jahn, D. | | Deposit date: | 2004-08-09 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tracking the Evolution of Porphobilinogen Synthase Metal Dependence in Vitro
J.Mol.Biol., 345, 2005
|
|
5WGT
 
 | | Crystal Structure of MalA' H253A, premalbrancheamide complex | | Descriptor: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
1W5T
 
 | | Structure of the Aeropyrum Pernix ORC2 protein (ADPNP-ADP complexes) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ORC2, ... | | Authors: | Singleton, M.R, Morales, R, Grainge, I, Cook, N, Isupov, M.N, Wigley, D.B. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes Induced by Nucleotide Binding in Cdc6/Orc from Aeropyrum Pernix
J.Mol.Biol., 343, 2004
|
|
7ORB
 
 | | Crystal structure of the L452R mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-75 and COVOX-253 Fabs | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
1W4J
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | PYRUVATE DEHYDROGENASE E2 | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-fast barrier-limited folding in the peripheral subunit-binding domain family.
J. Mol. Biol., 353, 2005
|
|
1W7E
 
 | |
1W4Y
 
 | | Ferrous horseradish peroxidase C1A in complex with carbon monoxide | | Descriptor: | CALCIUM ION, CARBON MONOXIDE, HORSERADISH PEROXIDASE C1A, ... | | Authors: | Carlsson, G.H, Nicholls, P, Svistunenko, D, Berglund, G.I, Hajdu, J. | | Deposit date: | 2004-08-03 | | Release date: | 2005-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complexes of Horseradish Peroxidase with Formate, Acetate, and Carbon Monoxide
Biochemistry, 44, 2005
|
|
4V7T
 
 | | Crystal structure of the E. coli ribosome bound to chloramphenicol. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.1942 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5WHR
 
 | | Discovery of a novel and selective IDO-1 inhibitor PF-06840003 and its characterization as a potential clinical candidate. | | Descriptor: | (3R)-3-(5-fluoro-1H-indol-3-yl)pyrrolidine-2,5-dione, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Greasley, S.E, Kaiser, S.E, Feng, J.L, Stewart, A. | | Deposit date: | 2017-07-18 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of a Novel and Selective Indoleamine 2,3-Dioxygenase (IDO-1) Inhibitor 3-(5-Fluoro-1H-indol-3-yl)pyrrolidine-2,5-dione (EOS200271/PF-06840003) and Its Characterization as a Potential Clinical Candidate.
J. Med. Chem., 60, 2017
|
|
1W8P
 
 | | Structural properties of the B25Tyr-NMe-B26Phe insulin mutant. | | Descriptor: | INSULIN A-CHAIN, INSULIN B-CHAIN, PHENOL, ... | | Authors: | Zakowa, L, Au-Alvarez, O, Dodson, E.J, Dodson, G.G, Brzozowski, A.M. | | Deposit date: | 2004-09-24 | | Release date: | 2005-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Towards the Insulin-Igf-I Intermediate Structures: Functional and Structural Properties of the B25Tyr-Nme-B26Phe Insulin Mutant.
Biochemistry, 43, 2004
|
|
4V8E
 
 | | Crystal structure analysis of ribosomal decoding (near-cognate tRNA-tyr complex). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
5W9J
 
 | |
1W91
 
 | |
1WAX
 
 | | Protein tyrosine phosphatase 1B with active site inhibitor | | Descriptor: | MAGNESIUM ION, PROTEIN-TYROSINE PHOSPHATASE, [[4-(AMINOMETHYL)PHENYL]AMINO]OXO-ACETIC ACID, | | Authors: | Hartshorn, M.J, Murray, C.W, Cleasby, A, Frederickson, M, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-10-28 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
1W7V
 
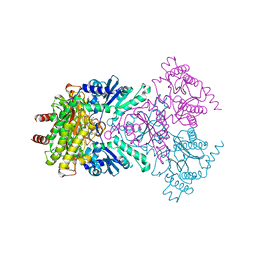 | | ZnMg substituted aminopeptidase P from E. coli | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PEPTIDE VAL-PRO-LEU, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-09-13 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Biochemistry, 44, 2005
|
|
6COC
 
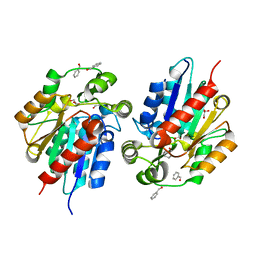 | | AtHNL enantioselectivity mutant At-A9-H7 Apo, Y13C,Y121L,P126F,L128W,C131T,F179L,A209I with benzaldehyde | | Descriptor: | Alpha-hydroxynitrile lyase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
4V9P
 
 | |
1W3D
 
 | |
5WB1
 
 | |
