5T1I
 
 | | CBX3 chromo shadow domain in complex with histone H3 peptide | | Descriptor: | Chromobox protein homolog 3, Histone H3.1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Peptide recognition by heterochromatin protein 1 (HP1) chromoshadow domains revisited: Plasticity in the pseudosymmetric histone binding site of human HP1.
J. Biol. Chem., 292, 2017
|
|
3EMH
 
 | | Structural basis of WDR5-MLL interaction | | Descriptor: | Mixed-lineage leukemia protein 1, SULFATE ION, WD repeat-containing protein 5 | | Authors: | Song, J.J, Kingston, R.E. | | Deposit date: | 2008-09-24 | | Release date: | 2008-10-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | WDR5 Interacts with Mixed Lineage Leukemia (MLL) Protein via the Histone H3-binding Pocket.
J.Biol.Chem., 283, 2008
|
|
2HUE
 
 | | Structure of the H3-H4 chaperone Asf1 bound to histones H3 and H4 | | Descriptor: | Anti-silencing protein 1, GLYCEROL, Histone H3, ... | | Authors: | English, C.M, Churchill, M.E.A, Tyler, J.K. | | Deposit date: | 2006-07-26 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the histone chaperone activity of asf1.
Cell(Cambridge,Mass.), 127, 2006
|
|
5AGC
 
 | | Crystallographic forms of the Vps75 tetramer | | Descriptor: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 75 | | Authors: | Hammond, C.M, Sundaramoorthy, R, Owen-Hughes, T. | | Deposit date: | 2015-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Histone Chaperone Vps75 Forms Multiple Oligomeric Assemblies Capable of Mediating Exchange between Histone H3-H4 Tetramers and Asf1-H3-H4 Complexes.
Nucleic Acids Res., 44, 2016
|
|
7QCE
 
 | | Crystal structure of an atypical PHD finger of VIN3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, VIN3 (Protein VERNALIZATION INSENSITIVE 3), ZINC ION | | Authors: | Franco-Echevarria, E, Fiedler, M, Dean, C, Bienz, M. | | Deposit date: | 2021-11-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plant vernalization proteins contain unusual PHD superdomains without histone H3 binding activity.
J.Biol.Chem., 298, 2022
|
|
6U04
 
 | |
2I7K
 
 | | Solution Structure of the Bromodomain of Human BRD7 Protein | | Descriptor: | Bromodomain-containing protein 7 | | Authors: | Sun, H, Liu, J, Zhang, J, Huang, H, Wu, J, Shi, Y. | | Deposit date: | 2006-08-31 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4
Biochem.Biophys.Res.Commun., 358, 2007
|
|
3TW1
 
 | | Structure of Rtt106-AHN | | Descriptor: | GLYCEROL, Histone chaperone RTT106, N-[2-(1H-IMIDAZOL-4-YL)ETHYL]ACETAMIDE | | Authors: | Su, D, Thompson, J.R, Mer, G. | | Deposit date: | 2011-09-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106.
Nature, 483, 2012
|
|
7CIZ
 
 | |
7CJ0
 
 | |
7OAQ
 
 | | Nanobody H3 AND C1 bound to RBD with Kent mutation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAP
 
 | | Nanobody H3 AND C1 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C1 nanobody, CHLORIDE ION, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
2FSA
 
 | |
5J3V
 
 | |
3TVV
 
 | |
6LKB
 
 | | Crystal Structure of the peptidylprolyl isomerase domain of Arabidopsis thaliana CYP71. | | Descriptor: | COBALT (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lakhanpal, S, Jobichen, C, Swaminathan, K. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and functional analyses of the PPIase domain of Arabidopsis thaliana CYP71 reveal its catalytic activity toward histone H3.
Febs Lett., 595, 2021
|
|
5VCH
 
 | |
4A7J
 
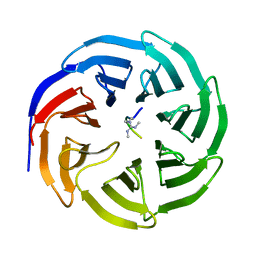 | | Symmetric Dimethylation of H3 Arginine 2 is a Novel Histone Mark that Supports Euchromatin Maintenance | | Descriptor: | HISTONE H3.1T, WD REPEAT-CONTAINING PROTEIN 5 | | Authors: | Migliori, V, Muller, J, Phalke, S, Low, D, Bezzi, M, ChuenMok, W, Gunaratne, J, Capasso, P, Bassi, C, Cecatiello, V, DeMarco, A, Blackstock, W, Kuznetsov, V, Amati, B, Mapelli, M, Guccione, E. | | Deposit date: | 2011-11-14 | | Release date: | 2012-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Dimethylation of H3R2 is a Newly Identified Histone Mark that Supports Euchromatin Maintenance
Nat.Struct.Mol.Biol., 19, 2012
|
|
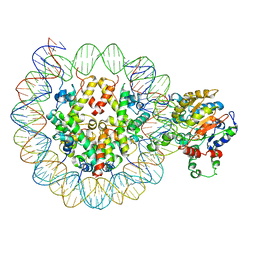
6PWF
 
 | |
5W0V
 
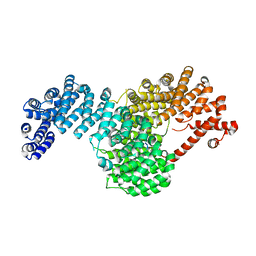 | | Crystal structure of full-length Kluyveromyces lactis Kap123 with histone H4 1-34 | | Descriptor: | Histone H4 1-34, Kap123 | | Authors: | An, S, Yoon, J, Song, J.-J, Cho, U.-S. | | Deposit date: | 2017-05-31 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | Structure-based nuclear import mechanism of histones H3 and H4 mediated by Kap123.
Elife, 6, 2017
|
|
2F6J
 
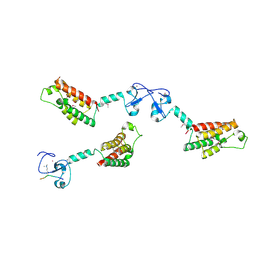 | |
6PWE
 
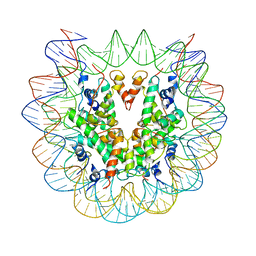 | | Cryo-EM structure of nucleosome core particle | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B, ... | | Authors: | Chittori, S, Subramaniam, S. | | Deposit date: | 2019-07-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structure of the primed state of the ATPase domain of chromatin remodeling factor ISWI bound to the nucleosome.
Nucleic Acids Res., 47, 2019
|
|
2JMI
 
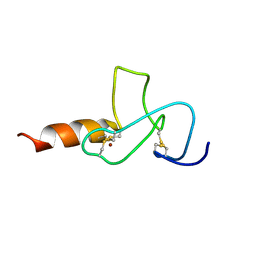 | | NMR solution structure of PHD finger fragment of Yeast Yng1 protein in free state | | Descriptor: | Protein YNG1, ZINC ION | | Authors: | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
4N4G
 
 | |
4OUC
 
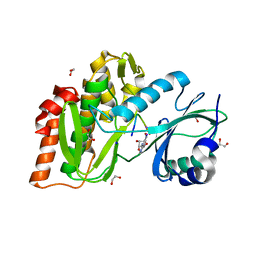 | | Structure of human haspin in complex with histone H3 substrate | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, Histone H3.2, ... | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-02-15 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of the chromatin phosphoproteome by the haspin protein kinase.
Mol Cell Proteomics, 13, 2014
|
|
