1PBV
 
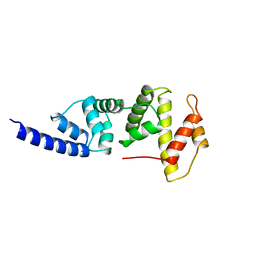 | | SEC7 DOMAIN OF THE EXCHANGE FACTOR ARNO | | Descriptor: | ARNO | | Authors: | Cherfils, J, Menetrey, J, Mathieu, M, Le Bras, G, Robineau, S, Beraud-Dufour, S, Antonny, B, Chardin, P. | | Deposit date: | 1998-01-15 | | Release date: | 1999-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Sec7 domain of the Arf exchange factor ARNO.
Nature, 392, 1998
|
|
1D9W
 
 | |
1BXP
 
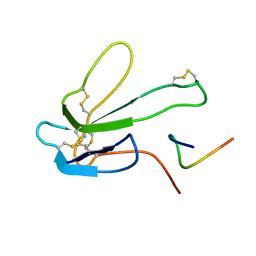 | | SOLUTION NMR STRUCTURE OF THE COMPLEX OF ALPHA-BUNGAROTOXIN WITH A LIBRARY DERIVED PEPTIDE, 20 STRUCTURES | | Descriptor: | ALPHA-BUNGAROTOXIN, PEPTIDE MET-ARG-TYR-TYR-GLU-SER-SER-LEU-LYS-SER-TYR-PRO-ASP | | Authors: | Scherf, T, Balass, M, Fuchs, S, Katchalski-Katzir, E, Anglister, J. | | Deposit date: | 1998-08-23 | | Release date: | 1999-01-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the complex of alpha-bungarotoxin with a library-derived peptide.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1NDC
 
 | |
4KIS
 
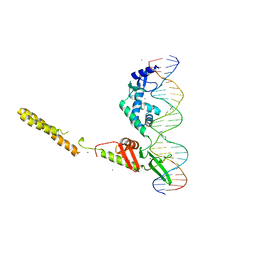 | | Crystal Structure of a LSR-DNA Complex | | Descriptor: | CALCIUM ION, DNA (26-MER), Putative integrase [Bacteriophage A118], ... | | Authors: | Rutherford, K, Yuan, P, Perry, K, Van Duyne, G.D. | | Deposit date: | 2013-05-02 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Attachment site recognition and regulation of directionality by the serine integrases.
Nucleic Acids Res., 41, 2013
|
|
5T3W
 
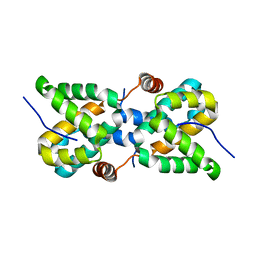 | |
5T3T
 
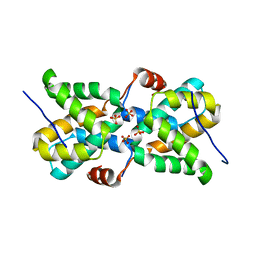 | | Ebola virus VP30 CTD bound to nucleoprotein | | Descriptor: | Fusion protein of Nucleoprotein and Minor nucleoprotein VP30, SULFATE ION | | Authors: | Kirchdoerfer, R.N, Moyer, C.L, Abelson, D.M, Saphire, E.O. | | Deposit date: | 2016-08-26 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Ebola Virus VP30-NP Interaction Is a Regulator of Viral RNA Synthesis.
Plos Pathog., 12, 2016
|
|
2RAP
 
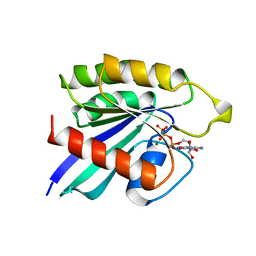 | | THE SMALL G PROTEIN RAP2A IN COMPLEX WITH GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RAP2A | | Authors: | Cherfils, J, Menetrey, J, Lebras, G. | | Deposit date: | 1998-05-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the small G protein Rap2A in complex with its substrate GTP, with GDP and with GTPgammaS.
EMBO J., 16, 1997
|
|
1B2S
 
 | |
1B3S
 
 | |
1B2U
 
 | |
3KFD
 
 | |
1B27
 
 | |
7E57
 
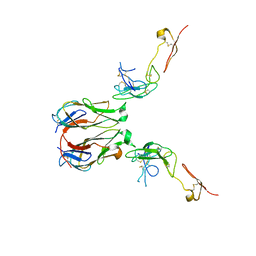 | | Crystal structure of murine GITR-GITRL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, Tumor necrosis factor receptor superfamily member 18, ... | | Authors: | Zhao, M, Tan, S, Fu, L, Chai, Y, Qi, J, Gao, G.F. | | Deposit date: | 2021-02-18 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Atypical TNF-TNFR superfamily binding interface in the GITR-GITRL complex for T cell activation.
Cell Rep, 36, 2021
|
|
1AXI
 
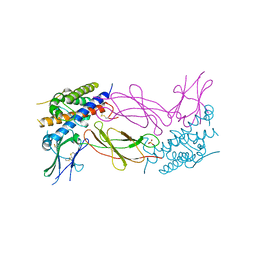 | | STRUCTURAL PLASTICITY AT THE HGH:HGHBP INTERFACE | | Descriptor: | GROWTH HORMONE, GROWTH HORMONE RECEPTOR, SULFATE ION | | Authors: | Atwell, S, Ultsch, M, De Vos, A.M, Wells, J.A. | | Deposit date: | 1997-10-15 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural plasticity in a remodeled protein-protein interface.
Science, 278, 1997
|
|
6GE6
 
 | |
6GE3
 
 | |
6R5L
 
 | | Fragment AZ-006 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-[[(2~{S})-1-azanylpropan-2-yl]amino]-6-(sulfanylmethyl)-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RK8
 
 | | Fragment AZ-014 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 7-(3-azanyl-4-methyl-pyrazol-1-yl)-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RL3
 
 | | Fragment AZ-003 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-azanyl-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RL6
 
 | | Fragment AZ-024 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Cellular tumor antigen p53, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RKK
 
 | | Fragment AZ-021 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-phenyl-5-(phenylmethyl)thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RJZ
 
 | | Fragment AZ-015 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 7-(6-azanylpyridin-2-yl)-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RX2
 
 | | Fragment AZ-005 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 7-[[(2~{S})-1-azanylpropan-2-yl]amino]-1-benzothiophene-2-carboximidamide, CALCIUM ION, ... | | Authors: | Leysen, S, Guillory, X, Wolter, M, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-06-07 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6S39
 
 | | Fragment AZ-018 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-(3-azanylpropyl)-4-phenyl-thiophene-2-carboximidamide, CALCIUM ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
