4K6W
 
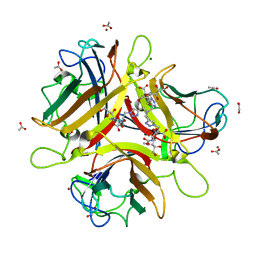 | | Crystal structure of Ad37 fiber knob in complex with trivalent sialic acid inhibitor ME0408 | | Descriptor: | 1,2-ETHANEDIOL, 2,2',2''-[nitrilotris(methanediyl-1H-1,2,3-triazole-4,1-diyl)]triethanol, 3,5-dideoxy-5-(propanoylamino)-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, ... | | Authors: | Stehle, T, Bauer, J. | | Deposit date: | 2013-04-16 | | Release date: | 2014-10-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Triazole linker-based trivalent sialic acid inhibitors of adenovirus type 37 infection of human corneal epithelial cells.
Org.Biomol.Chem., 13, 2015
|
|
7FQI
 
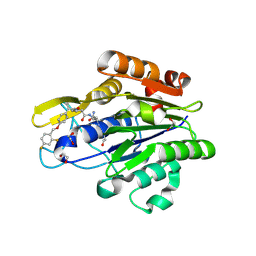 | | Crystal Structure of human Legumain in complex with (2S)-N-[(1S)-3-amino-1-cyano-3-oxopropyl]-1-[1-[4-[(2,4-difluorophenyl)methoxy]phenyl]cyclopropanecarbonyl]pyrrolidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Legumain, ... | | Authors: | Ehler, A, Benz, J, Bartels, B, Rudolph, M.G. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a human Legumain complex
To be published
|
|
7FQJ
 
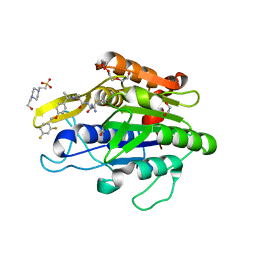 | | Crystal Structure of human Legumain in complex with (2S)-N-[(1S)-3-amino-1-cyano-3-oxopropyl]-1-[1-[4-[(2,4-difluorophenyl)methoxy]phenyl]cyclopropanecarbonyl]pyrrolidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ehler, A, Benz, J, Bartels, B, Rudolph, M.G. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a human Legumain complex
To be published
|
|
7AK5
 
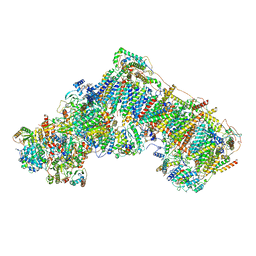 | | Cryo-EM structure of respiratory complex I in the deactive state from Mus musculus at 3.2 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yin, Z, Bridges, H.R, Grba, D, Hirst, J. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural basis for a complex I mutation that blocks pathological ROS production.
Nat Commun, 12, 2021
|
|
2W0V
 
 | | Crystal structure of Glmu from Haemophilus influenzae in complex with quinazoline inhibitor 1 | | Descriptor: | 6-(CYCLOPROP-2-EN-1-YLMETHOXY)-2-[6-(CYCLOPROPYLMETHYL)-5-OXO-3,4,5,6-TETRAHYDRO-2,6-NAPHTHYRIDIN-2(1H)-YL]-7-METHOXYQUINAZOLIN-4(3H)-ONE, GLUCOSAMINE-1-PHOSPHATE N-ACETYLTRANSFERASE, SULFATE ION, ... | | Authors: | Mochalkin, I, Melnick, M. | | Deposit date: | 2008-10-10 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery and Initial Sar of Quinazoline Inhibitors of Glmu from Haemophilus Influenzae
To be Published
|
|
5AB0
 
 | | Crystal structure of aminopeptidase ERAP2 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Giastas, P, Saridakis, E, Stratikos, E. | | Deposit date: | 2015-07-31 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (Er) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
3S97
 
 | | PTPRZ CNTN1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-1, Receptor-type tyrosine-protein phosphatase zeta | | Authors: | Bouyain, S. | | Deposit date: | 2011-05-31 | | Release date: | 2011-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2971 Å) | | Cite: | A complex between contactin-1 and the protein tyrosine phosphatase PTPRZ controls the development of oligodendrocyte precursor cells.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2NT7
 
 | | Crystal structure of PTP1B-inhibitor complex | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 1, {[5-(3-{[1-(BENZYLSULFONYL)PIPERIDIN-4-YL]AMINO}PHENYL)-4-BROMO-2-(2H-TETRAZOL-5-YL)-3-THIENYL]OXY}ACETIC ACID | | Authors: | Xu, W, Follows, B. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing acid replacements of thiophene PTP1B inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5P97
 
 | | rat catechol O-methyltransferase in complex with 5-(4-fluorophenyl)-2,3-dihydroxy-N-[(1-methylimidazol-4-yl)methyl]benzamide at 1.30A | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 5-(4-fluorophenyl)-2,3-dihydroxy-N-[(1-methylimidazol-4-yl)methyl]benzamide, CHLORIDE ION, ... | | Authors: | Ehler, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2016-08-30 | | Release date: | 2017-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of a COMT complex
To be published
|
|
1AAJ
 
 | |
7G4S
 
 | | Crystal Structure of rat Autotaxin in complex with 3-(4-chlorophenyl)-5-methyl-N-(oxolan-2-ylmethyl)pyrazolo[1,5-a]pyrimidin-7-amine, i.e. SMILES C12=C(C=NN1C(=CC(=N2)C)NC[C@@H]1OCCC1)c1ccc(cc1)Cl with IC50=0.95455 microM | | Descriptor: | (8R)-3-(4-chlorophenyl)-5-methyl-N-{[(2R)-oxolan-2-yl]methyl}pyrazolo[1,5-a]pyrimidin-7-amine, (8R)-3-(4-chlorophenyl)-5-methyl-N-{[(2S)-oxolan-2-yl]methyl}pyrazolo[1,5-a]pyrimidin-7-amine, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
1ZCK
 
 | | native structure prl-1 (ptp4a1) | | Descriptor: | ACETIC ACID, protein tyrosine phosphatase 4a1 | | Authors: | Sun, J.P, Wang, W.Q, Yang, H, Liu, S, Liang, F, Fedorov, A.A, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2005-04-12 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Biochemical Properties of PRL-1, a Phosphatase Implicated in Cell Growth, Differentiation, and Tumor Invasion.
Biochemistry, 44, 2005
|
|
7K6F
 
 | | Crystal structure of the tandem bromodomain (BD1, BD2) of human TAF1 in complex with MES (2-(N-morpholino)ethanesulfonic acid) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2020-09-20 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of Dual TAF1-ATR Inhibitors and Ligand-Induced Structural Changes of the TAF1 Tandem Bromodomain.
J.Med.Chem., 65, 2022
|
|
4PL9
 
 | |
1ONP
 
 | | IspC complex with Mn2+ and fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION | | Authors: | Steinbacher, S, Kaiser, J, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of fosmidomycin action revealed by the complex with 2-C-methyl-D-erythritol
4-phosphate synthase (IspC). Implications for the catalytic mechanism and anti-malaria
drug development.
J.BIOL.CHEM., 278, 2003
|
|
7K03
 
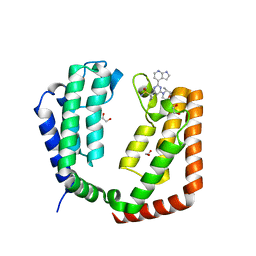 | |
7K1P
 
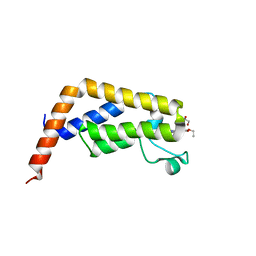 | |
3SHU
 
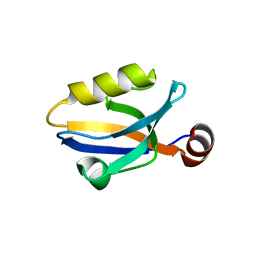 | | Crystal structure of ZO-1 PDZ3 | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Yu, J, Pan, L, Chen, J, Yu, H, Zhang, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Structure of the PDZ3-SH3-GuK Tandem of ZO-1 Suggests a Supramodular Organization of the Conserved MAGUK Family Scaffold Core
To be Published
|
|
1LPO
 
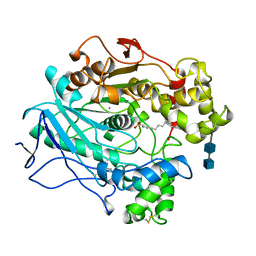 | |
4POZ
 
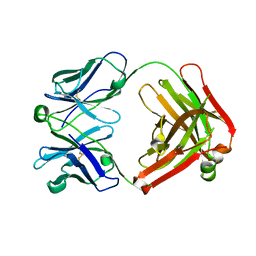 | | Fab fragment of Der p 1 specific antibody 10B9 | | Descriptor: | heavy chain of Fab fragment of 10B9 antibody, light chain of Fab fragment of 10B9 antibody | | Authors: | Osinski, T, Majorek, K.A, Pomes, A, Offermann, L.R, Osinski, S, Glesner, J, Vailes, L.D, Chapman, M.D, Minor, W, Chruszcz, M. | | Deposit date: | 2014-02-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Analysis of Der p 1-Antibody Complexes and Comparison with Complexes of Proteins or Peptides with Monoclonal Antibodies.
J. Immunol., 195, 2015
|
|
1LL3
 
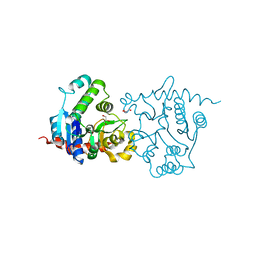 | |
7K42
 
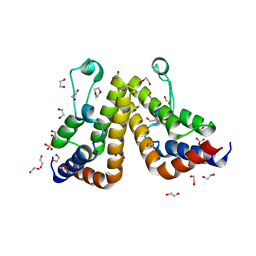 | | Crystal structure of the second bromodomain (BD2) of human TAF1 bound to dioxane | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, SULFATE ION, ... | | Authors: | Karim, M.R, Schonbrunn, E. | | Deposit date: | 2020-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Dual TAF1-ATR Inhibitors and Ligand-Induced Structural Changes of the TAF1 Tandem Bromodomain.
J.Med.Chem., 65, 2022
|
|
3ENZ
 
 | | Arsenolytic structure of Plasmodium falciparum purine nucleoside phosphorylase with hypoxanthine, ribose and arsenate ion | | Descriptor: | 1,4-anhydro-D-ribitol, ARSENATE, FORMIC ACID, ... | | Authors: | Chaikuad, A, Brady, R.L. | | Deposit date: | 2008-09-26 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Conservation of structure and activity in Plasmodium purine nucleoside phosphorylases.
Bmc Struct.Biol., 9, 2009
|
|
1UZU
 
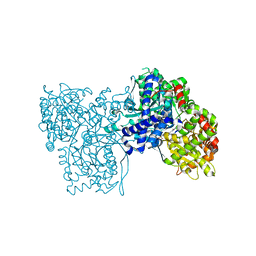 | | Glycogen Phosphorylase b in complex with indirubin-5'-sulphonate | | Descriptor: | 2',3-DIOXO-1,1',2',3-TETRAHYDRO-2,3'-BIINDOLE-5'-SULFONIC ACID, GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, ... | | Authors: | Kosmopoulou, M.N, Leonidas, D.D, Chrysina, E.D, Bischler, N, Eisenbrand, G, Sakarellos, C.E, Pauptit, R, Oikonomakos, N.G. | | Deposit date: | 2004-03-16 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of the potential antitumour agent indirubin-5-sulphonate at the inhibitor site of rabbit muscle glycogen phosphorylase b. Comparison with ligand binding to pCDK2-cyclin A complex.
Eur. J. Biochem., 271, 2004
|
|
3SL3
 
 | | Crystal structure of the apo form of the catalytic domain of PDE4D2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
