4DGL
 
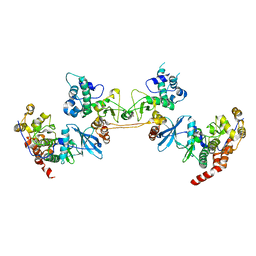 | | Crystal Structure of the CK2 Tetrameric Holoenzyme | | Descriptor: | Casein kinase II subunit alpha, Casein kinase II subunit beta, ZINC ION | | Authors: | Lolli, G, Battistutta, R. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural determinants of protein kinase CK2 regulation by autoinhibitory polymerization.
Acs Chem.Biol., 7, 2012
|
|
6MDZ
 
 | |
7B93
 
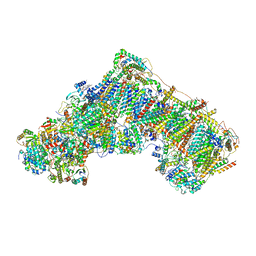 | | Cryo-EM structure of mitochondrial complex I from Mus musculus inhibited by IACS-2858 at 3.0 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1-[[3-(4-methylsulfonylpiperidin-1-yl)phenyl]methyl]-5-[3-[4-(trifluoromethyloxy)phenyl]-1,2,4-oxadiazol-5-yl]pyridin-2-one, ... | | Authors: | Chung, I, Hirst, J. | | Deposit date: | 2020-12-14 | | Release date: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Cork-in-bottle mechanism of inhibitor binding to mammalian complex I.
Sci Adv, 7, 2021
|
|
6LR4
 
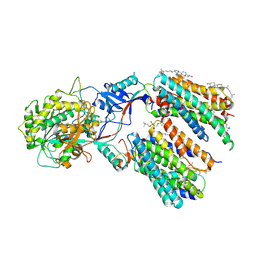 | | Molecular basis for inhibition of human gamma-secretase by small molecule | | Descriptor: | (2S)-2-hydroxy-3-methyl-N-[(2S)-1-[[(5S)-3-methyl-4-oxo-2,5-dihydro-1H-3-benzazepin-5-yl]amino]-1-oxopropan-2-yl]butanamide, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Shi, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
7W3P
 
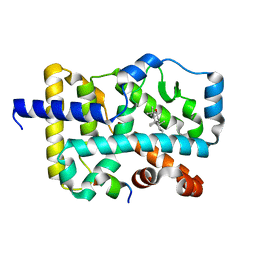 | | Crystal structure of RORgamma in complex with natural inverse agonist | | Descriptor: | (3S,5R,8R,9R,10R,12R,13R,14R,17S)-4,4,8,10,14-pentamethyl-17-[(2R)-2,6,6-trimethyloxan-2-yl]-2,3,5,6,7,9,11,12,13,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthrene-3,12-diol, Nuclear receptor ROR-gamma, Peptide from Nuclear receptor coactivator 2 | | Authors: | Tian, S.Y, Li, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of RORgamma in complex with natural inverse agonist
To Be Published
|
|
7BQ3
 
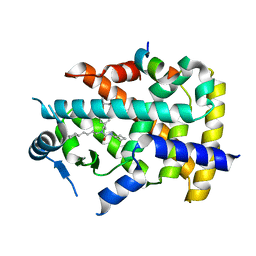 | | X-ray structure of human PPARalpha ligand binding domain-GW7647-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7RY5
 
 | | Cellular Retinoic Acid Binding Protein II with Bound Inhibitor 4-[6-({4-[(fluorosulfonyl)oxy]phenyl}ethynyl)-4,4-dimethyl-3,4-dihydroquinolin-1(2H)-yl]-4-oxobutanoic acid | | Descriptor: | 4-[6-({4-[(fluorosulfonyl)oxy]phenyl}ethynyl)-4,4-dimethyl-3,4-dihydroquinolin-1(2H)-yl]-4-oxobutanoic acid, Cellular retinoic acid-binding protein 2 | | Authors: | Kimmel, B.R, Mrksich, M. | | Deposit date: | 2021-08-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of an Enzyme-Inhibitor Reaction Using Cellular Retinoic Acid Binding Protein II for One-Pot Megamolecule Assembly.
Chemistry, 27, 2021
|
|
6S44
 
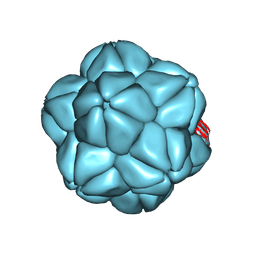 | |
3PGG
 
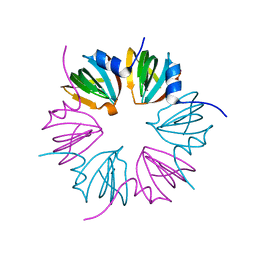 | | Crystal structure of cryptosporidium parvum u6 snrna-associated sm-like protein lsm5 | | Descriptor: | U6 snRNA-associated Sm-like protein LSm5. SM domain | | Authors: | Dong, A, Gao, M, Zhao, Y, Lew, J, Wasney, G.A, Kozieradzki, I, Vedadi, M, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-01 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Genome-Scale Protein Expression and Structural Biology of Plasmodium Falciparum and Related Apicomplexan Organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
7Y04
 
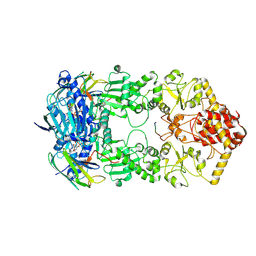 | | Hsp90-AhR-p23 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aryl hydrocarbon receptor, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-06-03 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
7SJ9
 
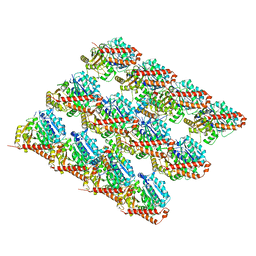 | | 13pf E254A microtubule from recombinant human tubulin decorated with EB3 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Microtubule-associated protein RP/EB family member 3, ... | | Authors: | LaFrance, B.J, Greber, B.J, Zhang, R, McCollum, C, Nogales, E. | | Deposit date: | 2021-10-16 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural transitions in the GTP cap visualized by cryo-electron microscopy of catalytically inactive microtubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6CBG
 
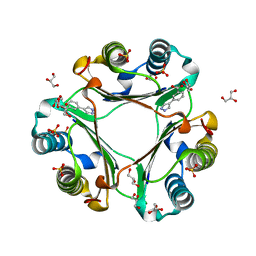 | | Macrophage Migration Inhibitory Factor in Complex with a Pyrazole Inhibitor (5) | | Descriptor: | 3-(1H-pyrazol-4-yl)benzoic acid, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Robertson, M.J, Krimmer, S.G, Jorgensen, W.L. | | Deposit date: | 2018-02-02 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of Pyrazoles as Phenol Surrogates to Yield Potent Inhibitors of Macrophage Migration Inhibitory Factor.
ChemMedChem, 13, 2018
|
|
6CIV
 
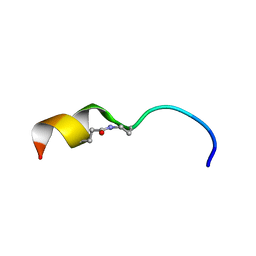 | |
5JX4
 
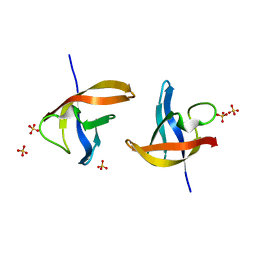 | | Crystal structure of E36-G37del mutant of the Bacillus caldolyticus cold shock protein. | | Descriptor: | Cold shock protein CspB, SULFATE ION | | Authors: | Carvajal, A, Castro-Fernandez, V, Cabrejos, D, Fuentealba, M, Pereira, H.M, Vallejos, G, Cabrera, R, Garratt, R.C, Komives, E.A, Ramirez-Sarmiento, C.A, Babul, J. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual dimerization of a BcCsp mutant leads to reduced conformational dynamics.
FEBS J., 284, 2017
|
|
3MUN
 
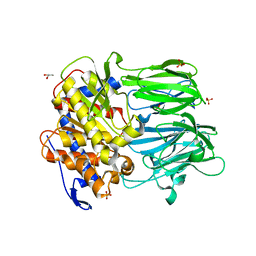 | | APPEP_PEPCLOSE closed state | | Descriptor: | GLYCEROL, Prolyl endopeptidase, SULFATE ION, ... | | Authors: | Chiu, T.K. | | Deposit date: | 2010-05-03 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Route of Substrate Entry in Prolyl Endopeptidase
TO BE PUBLISHED
|
|
8ONY
 
 | | Human Methionine Aminopeptidase 2 at the 80S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 60S ribosomal protein L19, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
7V3X
 
 | |
6FO8
 
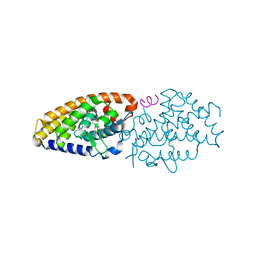 | | Vitamin D nuclear receptor complex 4 | | Descriptor: | (1~{R},3~{S},5~{Z})-4-methylidene-5-[(~{E})-3-[3-(6-methyl-6-oxidanyl-heptyl)phenyl]non-2-enylidene]cyclohexane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2018-02-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aromatic-Based Design of Highly Active and Noncalcemic Vitamin D Receptor Agonists.
J. Med. Chem., 61, 2018
|
|
3MUO
 
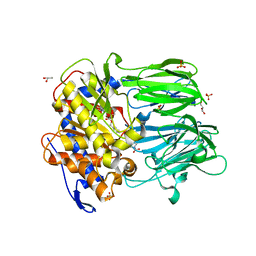 | | APPEP_PEPCLOSE+PP closed state | | Descriptor: | GLYCEROL, N-BENZYLOXYCARBONYL-L-PROLYL-L-PROLINAL, Prolyl endopeptidase, ... | | Authors: | Chiu, T.K. | | Deposit date: | 2010-05-03 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Route of Substrate Entry in Prolyl Endopeptidase
TO BE PUBLISHED
|
|
6N4O
 
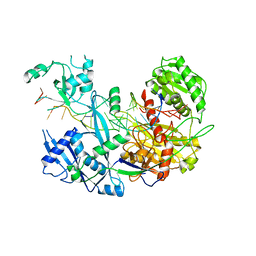 | |
6NBW
 
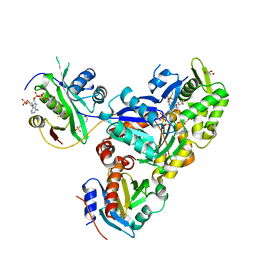 | | Ternary Complex of Beta/Gamma-Actin with Profilin and AnCoA-NAA80 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Rebowski, G, Boczkowska, M, Dominguez, R. | | Deposit date: | 2018-12-10 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of actin N-terminal acetylation.
Sci Adv, 6, 2020
|
|
6NDL
 
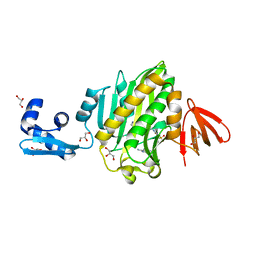 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with a sulfonamide inhibitor | | Descriptor: | 1-[4-(6-aminopurin-9-yl)butylsulfamoyl]-3-[4-[(4~{S})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butyl]urea, Biotin Protein Ligase, GLYCEROL | | Authors: | Marshall, A.C, Polyak, S.W, Bruning, J.B, Lee, K. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfonamide-Based Inhibitors of Biotin Protein Ligase as New Antibiotic Leads.
Acs Chem.Biol., 14, 2019
|
|
7E0A
 
 | | X-ray structure of human PPARgamma ligand binding domain-saroglitazar co-crystals obtained by co-crystallization | | Descriptor: | (2S)-2-ethoxy-3-[4-[2-[2-methyl-5-(4-methylsulfanylphenyl)pyrrol-1-yl]ethoxy]phenyl]propanoic acid, Isoform 2 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Uchii, K, Machida, Y, Oyama, T, Ishii, I. | | Deposit date: | 2021-01-27 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Structural Basis for Anti-non-alcoholic Fatty Liver Disease and Diabetic Dyslipidemia Drug Saroglitazar as a PPAR alpha / gamma Dual Agonist.
Biol.Pharm.Bull., 44, 2021
|
|
7E4W
 
 | |
6CBH
 
 | | Macrophage Migration Inhibitory Factor in Complex with a Pyrazole Inhibitor (8m) | | Descriptor: | 5-(3-fluoro-1H-pyrazol-4-yl)-2-[(naphthalen-2-yl)oxy]benzoic acid, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Robertson, M.J, Krimmer, S.G, Jorgensen, W.L. | | Deposit date: | 2018-02-02 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of Pyrazoles as Phenol Surrogates to Yield Potent Inhibitors of Macrophage Migration Inhibitory Factor.
ChemMedChem, 13, 2018
|
|
