7JO1
 
 | | Carbonic Anhydrase IX Mimic Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)butyramide | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)butanamide, ... | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
7JOB
 
 | | Carbonic Anhydrase IX Mimic Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)cyclohexanecarboxamide | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)cyclohexanecarboxamide, ... | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-06 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
1XIC
 
 | |
2P0W
 
 | | Human histone acetyltransferase 1 (HAT1) | | Descriptor: | ACETAMIDE, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Wu, H, Min, J, Zeng, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human histone acetyltransferase 1 (HAT1) in complex with acetylcoenzyme A and histone peptide H4
To be Published
|
|
1XL5
 
 | | HIV-1 Protease in complex with amidhyroxysulfone | | Descriptor: | CHLORIDE ION, N-{(1S)-1-(3-BROMOBENZYL)-4-[(4-BROMOPHENYL)SULFONYL]-6-METHYL-2-OXOHEPTYL}-2-(2,6-DIMETHYLPHENOXY)ACETAMIDE, PROTEASE RETROPEPSIN | | Authors: | Boettcher, J, Specker, E, Heine, A, Klebe, G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | An Old Target Revisited: Two New Privileged Skeletons and an Unexpected Binding Mode For HIV-Protease Inhibitors
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
7GSB
 
 | |
4JV3
 
 | | Crystal structure of beta-ketoacyl synthase from Brucella melitensis in complex with platencin | | Descriptor: | 2,4-dihydroxy-3-({3-[(2S,4aS,8S,8aR)-8-methyl-3-methylidene-7-oxo-1,3,4,7,8,8a-hexahydro-2H-2,4a-ethanonaphthalen-8-yl]propanoyl}amino)benzoic acid, Beta-ketoacyl synthase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of beta-ketoacyl ACP synthase I bound to platencin and fragment screening molecules at two substrate binding sites.
Proteins, 88, 2020
|
|
4HWV
 
 | |
3SWR
 
 | |
7KBR
 
 | | Co-crystal structure of alpha glucosidase with compound 10 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-nitro-4-(triazan-1-yl)phenyl]amino}ethyl (2-{[(1S,2S,3R,4S,5S)-2,3,4,5-tetrahydroxy-5-(hydroxymethyl)cyclohexyl]amino}ethyl)carbamate, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-10-02 | | Release date: | 2021-10-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
4E2W
 
 | | X-ray Structure of the H181N mutant of TcaB9, a C-3'-Methyltransferase, in Complex with S-Adenosyl-L-Homocysteine and Sugar Product | | Descriptor: | (2R,4S,6R)-4-amino-4,6-dimethyl-5-oxotetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Bruender, N.A, Holden, H.M. | | Deposit date: | 2012-03-09 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the catalytic mechanism of a C-3'-methyltransferase involved in the biosynthesis of D-tetronitrose.
Protein Sci., 21, 2012
|
|
2VYA
 
 | | Crystal Structure of fatty acid amide hydrolase conjugated with the drug-like inhibitor PF-750 | | Descriptor: | 4-(quinolin-3-ylmethyl)piperidine-1-carboxylic acid, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1, ... | | Authors: | Mileni, M, Johnson, D.S, Wang, Z, Everdeen, D.S, Liimatta, M, Pabst, B, Bhattacharya, K, Nugent, R.A, Kamtekar, S, Cravatt, B.F, Ahn, K, Stevens, R.C. | | Deposit date: | 2008-07-22 | | Release date: | 2008-09-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Guided Inhibitor Design for Human Faah by Interspecies Active Site Conversion.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3I3D
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE (M542A) IN COMPLEX WITH IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Dymianiw, D, Minhas, B, Huber, R.E. | | Deposit date: | 2009-06-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of Met-542 as a guide for the conformational changes of Phe-601 that occur during the reaction of β-galactosidase (Escherichia coli).
Biochem.Cell Biol., 88, 2010
|
|
2VU7
 
 | | Atomic resolution (1.08 A) structure of purified thaumatin I grown in sodium meso-tartrate at 4 C | | Descriptor: | 1,2-ETHANEDIOL, S,R MESO-TARTARIC ACID, Thaumatin-1 | | Authors: | Jakoncic, J, Asherie, N, Ginsberg, C. | | Deposit date: | 2008-05-21 | | Release date: | 2009-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Tartrate Chirality Determines Thaumatin Crystal Habit
Cryst.Growth Des., 9, 2009
|
|
4G2W
 
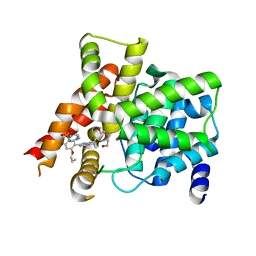 | | Crystal structure of PDE5A in complex with its inhibitor | | Descriptor: | 5,6-diethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Ren, J, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-07-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Design, synthesis, and pharmacological evaluation of monocyclic pyrimidinones as novel inhibitors of PDE5.
J.Med.Chem., 55, 2012
|
|
1VF4
 
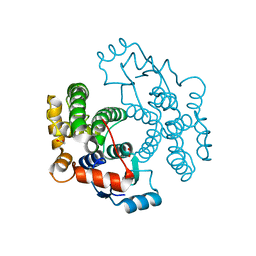 | | cGSTA1-1 apo form | | Descriptor: | ACETIC ACID, Glutathione S-transferase 3 | | Authors: | Lin, S.C, Lo, Y.C, Tam, M.F, Liaw, Y.C. | | Deposit date: | 2004-04-08 | | Release date: | 2005-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of chicken glutathione S-transferase A1-1
To be Published
|
|
1VST
 
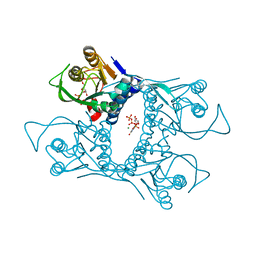 | | Symmetric Sulfolobus solfataricus uracil phosphoribosyltransferase with bound PRPP and GTP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kadziola, A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-09-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and kinetic studies of the allosteric transition in Sulfolobus solfataricus uracil phosphoribosyltransferase: Permanent activation by engineering of the C-terminus
J.Mol.Biol., 393, 2009
|
|
1I76
 
 | | COMPLEX OF 2-(BIPHENYL-4-SULFONYL)-1,2,3,4-TETRAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID (D-TIC DERIVATIVE) WITH T CATALITIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) | | Descriptor: | 2-(BIPHENYL-4-SULFONYL)-1,2,3,4-TETRAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID, CALCIUM ION, NEUTROPHIL COLLAGENASE, ... | | Authors: | Gavuzzo, E, Pochetti, G, Mazza, F, Gallina, C, Gorini, B, D'Alessio, S, Pieper, M, Tschesche, H, Tucker, P.A. | | Deposit date: | 2001-03-08 | | Release date: | 2001-03-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Two crystal structures of human neutrophil collagenase, one complexed with a primed- and the other with an unprimed-side inhibitor: implications for drug design.
J.Med.Chem., 43, 2000
|
|
5VZS
 
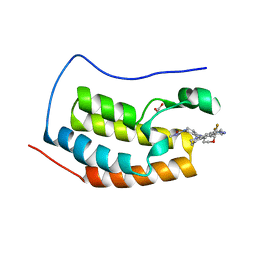 | | BRD4-BD1 in complex with Cpd19 (3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-N-methyl-1-(tetrahydro-2H-pyran-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide) | | Descriptor: | 1,2-ETHANEDIOL, 3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1-(oxan-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, Bromodomain-containing protein 4 | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-29 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of CBP/P300
To be published
|
|
5ZXG
 
 | | Cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from Arthrobacter globiformis, ligand-free form | | Descriptor: | CALCIUM ION, Cyclic maltosyl-maltose hydrolase | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2018-05-20 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural features of a bacterial cyclic alpha-maltosyl-(1→6)-maltose (CMM) hydrolase critical for CMM recognition and hydrolysis.
J. Biol. Chem., 293, 2018
|
|
2OXF
 
 | | X-ray structure of the unliganded uridine phosphorylase from Salmonella typhimurium in homodimeric form at 1.76A resolution | | Descriptor: | GLYCEROL, Uridine phosphorylase | | Authors: | Timofeev, V.I, Pavlyk, B.P, Lashkov, A.A, Gabdoulkhakov, A.G, Mikhailov, A.M. | | Deposit date: | 2007-02-20 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray structure of the unliganded uridine phosphorylase from Salmonella typhimurium in homodimeric form at 1.76A resolution
To be Published
|
|
1SQZ
 
 | | Design of specific inhibitors of Phopholipase A2: Crystal structure of the complex formed between Group II Phopholipase A2 and a designed peptide Dehydro-Ile-Ala-Arg-Ser at 1.2A resolution | | Descriptor: | Phospholipase A2, SULFATE ION, synthetic peptide | | Authors: | Singh, N, Prem Kumar, R, Somvanshi, R.K, Bilgrami, S, Ethayathulla, A.S, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2004-03-22 | | Release date: | 2004-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Design of specific inhibitors of Phopholipase A2: Crystal structure of the complex formed between GroupII Phopholipase A2 and a designed peptide Dehydro-Ile-Ala-Arg-Ser at 1.2A resolution
To be Published
|
|
2VTF
 
 | | X-ray crystal structure of the Endo-beta-N-acetylglucosaminidase from Arthrobacter protophormiae E173Q mutant reveals a TIM barrel catalytic domain and two ancillary domains | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-BETA-N-ACETYLGLUCOSAMINIDASE, TRIETHYLENE GLYCOL | | Authors: | Ling, Z, Bingham, R.J, Suits, M.D.L, Moir, J.W.B, Fairbanks, A.J, Taylor, E.J. | | Deposit date: | 2008-05-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The X-Ray Crystal Structure of an Arthrobacter Protophormiae Endo-Beta-N-Acetylglucosaminidase Reveals a (Beta/Alpha)(8) Catalytic Domain, Two Ancillary Domains and Active Site Residues Key for Transglycosylation Activity.
J.Mol.Biol., 389, 2009
|
|
6HA9
 
 | |
1AMC
 
 | |
