5VUJ
 
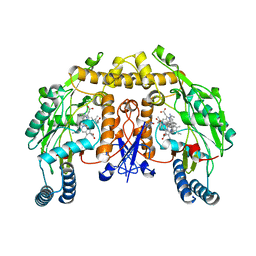 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(((3-(Dimethylamino)benzyl)amino)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[[3-(dimethylamino)phenyl]methylamino]methyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
5VUU
 
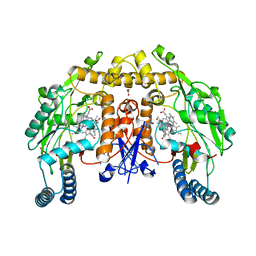 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 4-(2-(((2-Amino-4-methylquinolin-7-yl)methyl)amino)ethyl)-2-methylbenzonitrile | | Descriptor: | 4-(2-{[(2-amino-4-methylquinolin-7-yl)methyl]amino}ethyl)-2-methylbenzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
1VEW
 
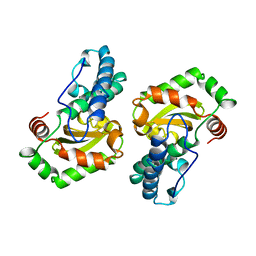 | | MANGANESE SUPEROXIDE DISMUTASE FROM ESCHERICHIA COLI | | Descriptor: | HYDROXIDE ION, MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Baker, H.M, Whittaker, M.M, Whittaker, J.W, Jameson, G.B, Baker, E.N. | | Deposit date: | 1998-01-20 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Escherichia coli manganese superoxide dismutase at 2.1-angstrom resolution.
J.Biol.Inorg.Chem., 3, 1998
|
|
1VFI
 
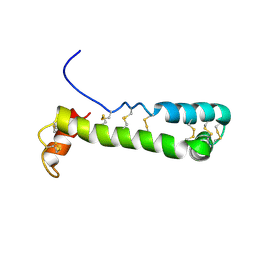 | | Solution Structure of Vanabin2 (RUH-017), a Vanadium-binding Protein from Ascidia sydneiensis samea | | Descriptor: | vanadium-binding protein 2 | | Authors: | Hamada, T, Hirota, H, Asanuma, M, Hayashi, F, Kobayashi, N, Ueki, T, Michibata, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-04-13 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Vanabin2, a Vanadium(IV)-Binding Protein from the Vanadium-Rich Ascidian Ascidia sydneiensis samea
J.Am.Chem.Soc., 127, 2005
|
|
5W3T
 
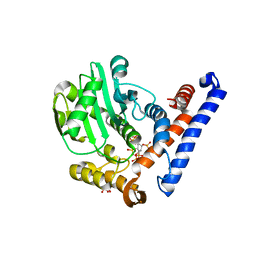 | | Crystal structure of PopP2 in complex with IP6 | | Descriptor: | GLYCEROL, INOSITOL HEXAKISPHOSPHATE, PopP2 protein | | Authors: | Song, J, Zhang, Z.M. | | Deposit date: | 2017-06-08 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanism of host substrate acetylation by a YopJ family effector.
Nat Plants, 3, 2017
|
|
5VV5
 
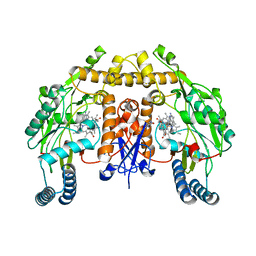 | | Structure of human neuronal nitric oxide synthase heme domain in complex with 4-(2-(((2-Amino-4-methylquinolin-7-yl)methyl)amino)ethyl)-2-methylbenzonitrile | | Descriptor: | 4-(2-{[(2-amino-4-methylquinolin-7-yl)methyl]amino}ethyl)-2-methylbenzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
5VW5
 
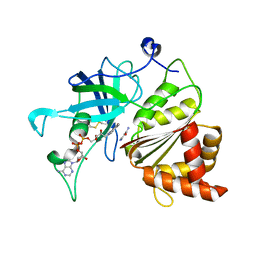 | | Nicotinamide soak of Y316A mutant of corn root ferredoxin:NADP+ reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, MAGNESIUM ION, ... | | Authors: | Kean, K.M, Carpenter, R.A, Hall, A.R, Karplus, P.A. | | Deposit date: | 2017-05-21 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | High-resolution studies of hydride transfer in the ferredoxin:NADP(+) reductase superfamily.
FEBS J., 284, 2017
|
|
1VHN
 
 | |
5W4C
 
 | | Crystal structure of thioredoxin reductase from Cryptococcus neoformans in complex with FAD (FO conformation) | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bravo-Chaucanes, C.P, Valadares, N.F, Felipe, M.S.S, Barbosa, J.A.R.G. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Crystal structure of thioredoxin reductase from Cryptococcus neoformans in complex with FAD (FO conformation)
To be published
|
|
5W4Z
 
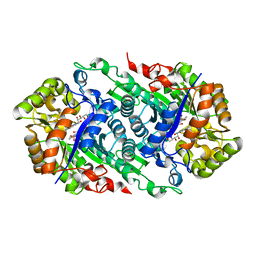 | | Crystal Structure of Riboflavin Lyase (RcaE) with modified FMN and substrate Riboflavin | | Descriptor: | 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)-3-O-phosphono-D-ribitol, RIBOFLAVIN, Riboflavin Lyase | | Authors: | Bhandari, D.M, Chakrabarty, Y, Zhao, B, Wood, J, Li, P, Begley, T.P. | | Deposit date: | 2017-06-13 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cannibalism Among the Flavins: a Novel C-N Bond Cleavage in Riboflavin Catabolism Mediated by Flavin-Generated Superoxide Radical
To be Published
|
|
1VJB
 
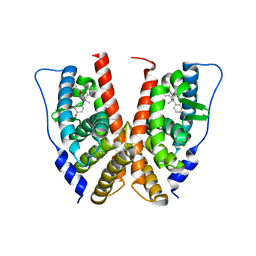 | | crystal structure of the ligand-binding domain of the estrogen-related receptor gamma in complex with 4-hydroxytamoxifen | | Descriptor: | 4-HYDROXYTAMOXIFEN, Estrogen-related receptor gamma | | Authors: | Greschik, H, Flaig, R, Renaud, J.P, Moras, D. | | Deposit date: | 2004-02-03 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for the Deactivation of the Estrogen-related Receptor {gamma} by Diethylstilbestrol or 4-Hydroxytamoxifen and Determinants of Selectivity.
J.Biol.Chem., 279, 2004
|
|
1VII
 
 | |
5M2H
 
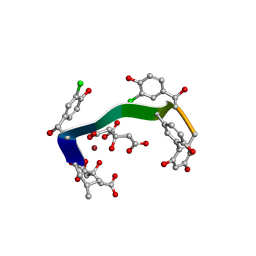 | | Crystal structure of vancomycin-Zn(II)-citrate complex | | Descriptor: | CITRIC ACID, HEXAETHYLENE GLYCOL, Vancomycin, ... | | Authors: | Zarkan, A, Macklyne, H.-R, Chirgadze, D.Y, Bond, A.D, Hesketh, A.R, Hong, H.-J. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Zn(II) mediates vancomycin polymerization and potentiates its antibiotic activity against resistant bacteria.
Sci Rep, 7, 2017
|
|
5W5P
 
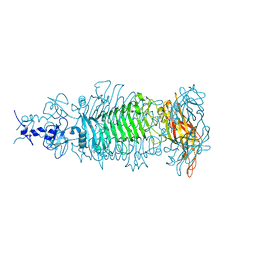 | |
1VPU
 
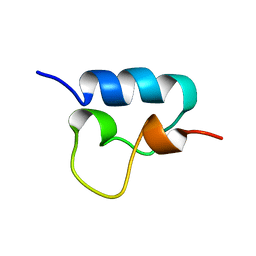 | |
5W5X
 
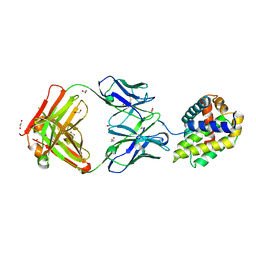 | | Crystal structure of BAXP168G in complex with an activating antibody | | Descriptor: | 1,2-ETHANEDIOL, 3C10 Fab' heavy chain, 3C10 Fab' light chain, ... | | Authors: | Robin, A.Y, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Ensemble Properties of Bax Determine Its Function.
Structure, 26, 2018
|
|
1VJQ
 
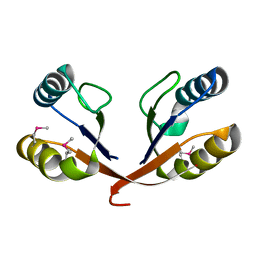 | |
5VYQ
 
 | | Crystal structure of the N-formyltransferase Rv3404c from mycobacterium tuberculosis in complex with YDP-Qui4N and folinic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dunsirn, M.M, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical Investigation of Rv3404c from Mycobacterium tuberculosis.
Biochemistry, 56, 2017
|
|
1VKH
 
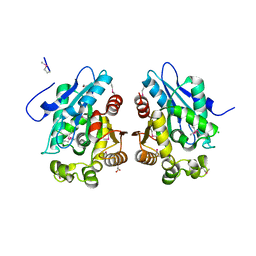 | |
1VCQ
 
 | |
1VCY
 
 | | VVA2 isoform | | Descriptor: | MALONATE ION, volvatoxin A2 | | Authors: | Lin, S.-C, Lo, Y.-C, Lin, J.-Y, Liaw, Y.-C. | | Deposit date: | 2004-03-17 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures and electron micrographs of fungal volvatoxin A2
J.Mol.Biol., 343, 2004
|
|
5W67
 
 | | HLA-C*06:02 presenting VRSRR(ABA)LRL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mobbs, J.I, Vivian, J.P, Rossjohn, J. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular basis for peptide repertoire selection in the human leucocyte antigen (HLA) C*06:02 molecule.
J. Biol. Chem., 292, 2017
|
|
5VZB
 
 | | Post-catalytic complex of human Polymerase Mu (G433S) mutant with incoming UTP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
1VD7
 
 | | Solution structure of FMBP-1 tandem repeat 1 | | Descriptor: | Fibroin-modulator-binding-protein-1 | | Authors: | Kawaguchi, K, Yamaki, T, Aizawa, T, Takiya, S, Demura, M, Nitta, K. | | Deposit date: | 2004-03-20 | | Release date: | 2005-03-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FMBP-1 tandem repeat 1
To be Published
|
|
1VDF
 
 | |
