3LNV
 
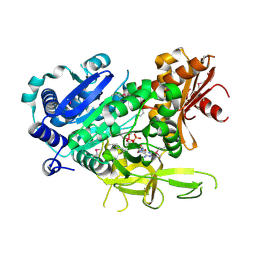 | | The crystal structure of fatty acyl-adenylate ligase from L. pneumophila in complex with acyl adenylate and pyrophosphate | | Descriptor: | 5'-O-[(S)-(dodecanoyloxy)(hydroxy)phosphoryl]adenosine, PYROPHOSPHATE 2-, Saframycin Mx1 synthetase B | | Authors: | Zhang, Z, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-03 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Studies of Fatty Acyl Adenylate Ligases from E. coli and L. pneumophila.
J.Mol.Biol., 406, 2011
|
|
8GRJ
 
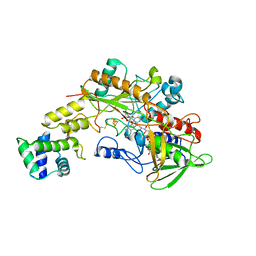 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yoshida, H, Kojima, K, Tsugawa, W, Okuda-Shimazaki, J, Kerrigan, J.A, Sode, K. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Improvement of substrate specificity of the direct electron transfer type FAD-dependent glucose dehydrogenase catalytic subunit.
J.Biotechnol., 2024
|
|
8EI4
 
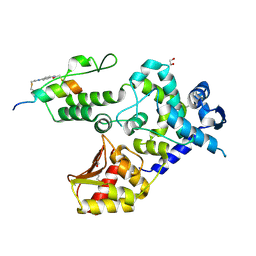 | | Crystal structure of the WWP1 HECT domain in complex with H302, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, H302, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
6VQN
 
 | | Co-crystal structure of human PD-L1 complexed with Compound A | | Descriptor: | N,N'-(2,2'-dimethyl[1,1'-biphenyl]-3,3'-diyl)bis(5-{[(2-hydroxyethyl)amino]methyl}pyridine-2-carboxamide), Programmed cell death 1 ligand 1 | | Authors: | White, A, Lakshminarasimhan, D, Leo, C, Suto, R.K. | | Deposit date: | 2020-02-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Checkpoint inhibition through small molecule-induced internalization of programmed death-ligand 1.
Nat Commun, 12, 2021
|
|
4NPQ
 
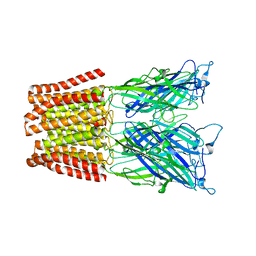 | | The resting-state conformation of the GLIC ligand-gated ion channel | | Descriptor: | Proton-gated ion channel | | Authors: | Sauguet, L, Shahsavar, A, Poitevin, F, Huon, C, Menny, A, Nemecz, A, Haouz, A, Changeux, J.P, Corringer, P.J, Delarue, M. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7Y3A
 
 | | Crystal structure of TRIM7 bound to 2C | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-2C | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8OHD
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ5
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (in-vitro reconstitution) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Peter, J.J, Kulathu, Y, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ0
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 2 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
4EGP
 
 | | The X-ray crystal structure of CYP199A4 in complex with 2-naphthoic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
6QLY
 
 | | IDOL FERM domain | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase MYLIP, SULFATE ION | | Authors: | Martinelli, L, Sixma, T.K. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
5C7J
 
 | | CRYSTAL STRUCTURE OF NEDD4 WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
1SLD
 
 | |
1C16
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE GAMMA/DELTA T CELL LIGAND T22 | | Descriptor: | MHC-LIKE PROTEIN T22, PROTEIN (BETA-2-MICROGLOBULIN) | | Authors: | Wingren, C, Crowley, M.P, Degano, M, Chien, Y, Wilson, I.A. | | Deposit date: | 1999-07-20 | | Release date: | 2000-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a gammadelta T cell receptor ligand T22: a truncated MHC-like fold.
Science, 287, 2000
|
|
3PT3
 
 | | Crystal structure of the C-terminal lobe of the human UBR5 HECT domain | | Descriptor: | E3 ubiquitin-protein ligase UBR5 | | Authors: | Matta-Camacho, E, Kozlov, G, Menade, M, Gehring, K. | | Deposit date: | 2010-12-02 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the HECT C-lobe of the UBR5 E3 ubiquitin ligase.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2A0Z
 
 | | The molecular structure of toll-like receptor 3 ligand binding domain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bell, J.K, Botos, I, Hall, P.R, Askins, J, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2005-06-17 | | Release date: | 2005-08-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular structure of the Toll-like receptor 3 ligand-binding domain
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1YIM
 
 | |
2P1Q
 
 | | Mechanism of Auxin Perception by the TIR1 ubiquitin ligase | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Auxin-responsive protein IAA7, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, N. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase.
Nature, 446, 2007
|
|
1YIN
 
 | |
2ZFO
 
 | | Structure of the partially unliganded met state of 400 kDa hemoglobin: Insights into ligand-induced structural changes of giant hemoglobins | | Descriptor: | Extracellular giant hemoglobin major globin subunit A1, Extracellular giant hemoglobin major globin subunit A2, Extracellular giant hemoglobin major globin subunit B1, ... | | Authors: | Numoto, N, Nakagawa, T, Kita, A, Sasayama, Y, Fukumori, Y, Miki, K. | | Deposit date: | 2008-01-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the partially unliganded met state of 400 kDa hemoglobin: insights into ligand-induced structural changes of giant hemoglobins
Proteins, 73, 2008
|
|
3PZC
 
 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with Coenzyme A | | Descriptor: | ACETATE ION, Amino acid--[acyl-carrier-protein] ligase 1, COENZYME A, ... | | Authors: | Weygand-Durasevic, I, Luic, M, Mocibob, M, Ivic, N, Subasic, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate Recognition by Novel Family of Amino Acid:[Carrier Protein] Ligases
Croatica Chemica Acta, 84, 2011
|
|
2UWX
 
 | | Active site restructuring regulates ligand recognition in class A penicillin-binding proteins | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Macheboeuf, P, DiGuilmi, A.M, Job, V, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2007-03-23 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Active Site Restructuring Regulates Ligand Recognition in Class a Penicillin-Binding Proteins
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
7Q8T
 
 | | Crystal structure of NAMPT bound to ligand TSY535(compound 9a) | | Descriptor: | Nicotinamide phosphoribosyltransferase, SULFATE ION, [(2~{R},3~{S},4~{R},5~{S})-3,4-bis(oxidanyl)-5-[4-[[[4-(phenylsulfonyl)phenyl]carbamoylamino]methyl]phenyl]oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Kraemer, A, Tang, S, Butterworth, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Chemistry-led investigations into the mode of action of NAMPT activators, resulting in the discovery of non-pyridyl class NAMPT activators.
Acta Pharm Sin B, 13, 2023
|
|
4F52
 
 | | Structure of a Glomulin-RBX1-CUL1 complex | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase RBX1, Glomulin, ... | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2012-05-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a Glomulin-RBX1-CUL1 Complex: Inhibition of a RING E3 Ligase through Masking of Its E2-Binding Surface.
Mol.Cell, 47, 2012
|
|
5HCM
 
 | |
