7VQJ
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed 1u. | | Descriptor: | (2R)-2-[(1S)-1-methoxy-2-oxidanyl-2-oxidanylidene-ethyl]-5-methylidene-2H-1,3-thiazine-4-carboxylic acid, Metallo-beta-lactamase NDM-1, ZINC ION | | Authors: | Xie, H.X. | | Deposit date: | 2021-10-20 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of NDM-1 in complex with hydrolyzed 1u.
To Be Published
|
|
5AKH
 
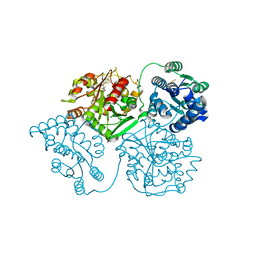 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 4-[5-[3-(trifluoromethyl)phenyl]-1H-pyrazol-4-yl]pyridine, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-03 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
7DHW
 
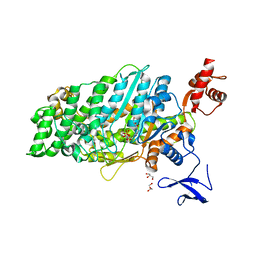 | | Crystal structure of myosin-XI motor domain in complex with ADP-ALF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Haraguchi, T, Tamanaha, M, Yoshimura, K, Imi, T, Tominaga, M, Sakayama, H, Nishiyama, T, Ito, K, Murata, T. | | Deposit date: | 2020-11-17 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of ultrafast myosin, its amino acid sequence, and structural features.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6BC9
 
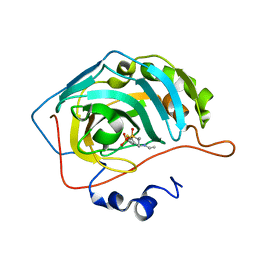 | | Joint X-ray/neutron structure of human carbonic anhydrase II in complex with dorzolamide | | Descriptor: | (4S-TRANS)-4-(ETHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Kovalevsky, A, McKenna, R, Aggarwal, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | "To Be or Not to Be" Protonated: Atomic Details of Human Carbonic Anhydrase-Clinical Drug Complexes by Neutron Crystallography and Simulation.
Structure, 26, 2018
|
|
7D4L
 
 | |
7OU0
 
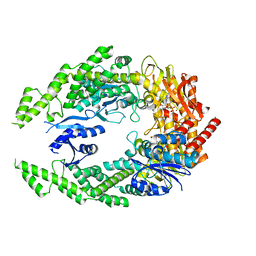 | | The structure of MutS bound to two molecules of ADP-Vanadate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS, MAGNESIUM ION, ... | | Authors: | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6B9G
 
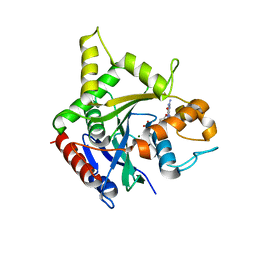 | | human ATL1 GTPase domain bound to GDP | | Descriptor: | Atlastin-1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | O'Donnell, J.P, Sondermann, H. | | Deposit date: | 2017-10-10 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A hereditary spastic paraplegia-associated atlastin variant exhibits defective allosteric coupling in the catalytic core.
J. Biol. Chem., 293, 2018
|
|
5AI4
 
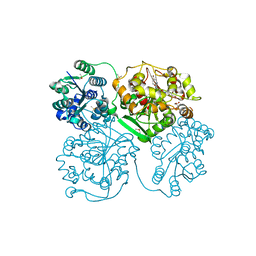 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 2-[(5-BROMO-2-PYRIDYL)-METHYL-AMINO]ETHANOL, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-02-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
7D4X
 
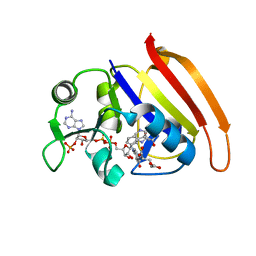 | |
7OU2
 
 | | The structure of MutS bound to two molecules of ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS | | Authors: | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | Deposit date: | 2021-06-11 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7K21
 
 | |
5ALF
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 2-phenyl-N,N-dipropyl-1H-benzimidazole-5-sulfonamide, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5ALL
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | DIMETHYL SULFOXIDE, SOLUBLE EPOXIDE HYDROLASE, SULFATE ION, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
6BMB
 
 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase (T381G mutant) in complex with tartrate and shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic, ... | | Authors: | Christendat, D, Peek, J. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | Structural and biochemical approaches uncover multiple evolutionary trajectories of plant quinate dehydrogenases.
Plant J., 2018
|
|
8CAL
 
 | |
8CM7
 
 | | W-formate dehydrogenase M405A from Desulfovibrio vulgaris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Vilela-Alves, G, Mota, C, Oliveira, A.R, Manuel, R.R, Pereira, I.C, Romao, M.J. | | Deposit date: | 2023-02-17 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | An allosteric redox switch involved in oxygen protection in a CO 2 reductase.
Nat.Chem.Biol., 20, 2024
|
|
7OY6
 
 | | Crystal structure of human DYRK1A in complex with ARN25068 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ~{N}4-(3-cyclopropyl-1~{H}-pyrazol-5-yl)-~{N}2-(phenylmethyl)thieno[3,2-d]pyrimidine-2,4-diamine | | Authors: | Tripathi, S.K, Balboni, B, Demuro, S, DiMartino, R, Ortega, J, Girotto, S, Cavalli, A. | | Deposit date: | 2021-06-23 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | ARN25068, a versatile starting point towards triple GSK-3 beta /FYN/DYRK1A inhibitors to tackle tau-related neurological disorders.
Eur.J.Med.Chem., 229, 2022
|
|
521P
 
 | | THREE-DIMENSIONAL STRUCTURES OF H-RAS P21 MUTANTS: MOLECULAR BASIS FOR THEIR INABILITY TO FUNCTION AS SIGNAL SWITCH MOLECULES | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, H-RAS P21 PROTEIN, MAGNESIUM ION | | Authors: | Schlichting, I, Krengel, U, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules.
Cell(Cambridge,Mass.), 62, 1990
|
|
7DPJ
 
 | | H-Ras Q61L in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Taniguchi, H, Matsumoto, S, Miyamoto, R, Kawamura, T, Kumasaka, T, Kataoka, T. | | Deposit date: | 2020-12-19 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.976 Å) | | Cite: | Oncogenic mutations Q61L and Q61H confer active form-like structural features to the inactive state (state 1) conformation of H-Ras protein.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
7DPH
 
 | | H-Ras Q61H in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Taniguchi, H, Matsumoto, S, Kawamura, T, Kumasaka, T, Kataoka, T. | | Deposit date: | 2020-12-19 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Oncogenic mutations Q61L and Q61H confer active form-like structural features to the inactive state (state 1) conformation of H-Ras protein.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
8CJ7
 
 | | HDAC6 selective degraded (difluoromethyl)-1,3,4-oxadiazole substrate inhibitor | | Descriptor: | 6-[(5-pyridin-2-yl-1,2$l^{4},3,4-tetrazacyclopenta-1,3-dien-2-yl)methyl]pyridine-3-carbohydrazide, Histone deacetylase 6, IODIDE ION, ... | | Authors: | Sandmark, J, Ek, M, Ripa, L. | | Deposit date: | 2023-02-12 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Selective and Bioavailable HDAC6 2-(Difluoromethyl)-1,3,4-oxadiazole Substrate Inhibitors and Modeling of Their Bioactivation Mechanism.
J.Med.Chem., 66, 2023
|
|
4ZX0
 
 | | Human Carbonic Anhydrase II in complex with a glucosyl sulfamate inhibitor | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, ZINC ION, ... | | Authors: | Mahon, B.P, Lomelino, C.L, Pinard, M.A, McKenna, R. | | Deposit date: | 2015-05-19 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping Selective Inhibition of the Cancer-Related Carbonic Anhydrase IX using Structure-Activity Relationships of Glucosyl-Based Sulfamates
J. Med. Chem., 58, 2015
|
|
7K13
 
 | | ACMSD in complex with diflunisal derivative 14 | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, 2-hydroxy-5-(thiophen-3-yl)benzoic acid, ZINC ION | | Authors: | Yang, Y, Liu, A. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Diflunisal Derivatives as Modulators of ACMS Decarboxylase Targeting the Tryptophan-Kynurenine Pathway.
J.Med.Chem., 64, 2021
|
|
6BMQ
 
 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase (T381G mutant) in complex with tartrate and shikimate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic, ... | | Authors: | Christendat, D, Peek, J. | | Deposit date: | 2017-11-15 | | Release date: | 2018-09-26 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | Structural and biochemical approaches uncover multiple evolutionary trajectories of plant quinate dehydrogenases.
Plant J., 2018
|
|
4ZZN
 
 | | Human ERK2 in complex with an inhibitor | | Descriptor: | 2-[[5-chloranyl-2-(oxan-4-ylamino)pyridin-4-yl]amino]-N-methyl-benzamide, MITOGEN-ACTIVATED PROTEIN KINASE 1, SULFATE ION | | Authors: | Ward, R.A, Colclough, N, Challinor, M, Debreczeni, J.E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, James, M, Jones, C.D, Jones, C.R, Renshaw, J, Roberts, K, Snow, L, Tonge, M, Yeung, K. | | Deposit date: | 2015-04-10 | | Release date: | 2015-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Guided Design of Highly Selective and Potent Covalent Inhibitors of Erk1/2.
J.Med.Chem., 58, 2015
|
|
