4X1F
 
 | | Crystal structure of the hPXR-LBD in complex with the synthetic estrogen 17alpha-ethinylestradiol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Ethinyl estradiol, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Delfosse, V, Huet, T, Bourguet, W. | | Deposit date: | 2014-11-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synergistic activation of human pregnane X receptor by binary cocktails of pharmaceutical and environmental compounds.
Nat Commun, 6, 2015
|
|
3HSA
 
 | |
3US9
 
 | | Crystal Structure of the NCX1 Intracellular Tandem Calcium Binding Domains(CBD12) | | Descriptor: | CALCIUM ION, Sodium/calcium exchanger 1 | | Authors: | Giladi, M, Sasson, Y, Hirsch, J.A, Khananshvili, D. | | Deposit date: | 2011-11-23 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A common Ca2+-driven interdomain module governs eukaryotic NCX regulation.
Plos One, 7, 2012
|
|
3O85
 
 | | Giardia lamblia 15.5kD RNA binding protein | | Descriptor: | Ribosomal protein L7Ae | | Authors: | Biswas, S, Maxwell, E.S. | | Deposit date: | 2010-08-02 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Comparative analysis of the 15.5kD box C/D snoRNP core protein in the primitive eukaryote Giardia lamblia reveals unique structural and functional features.
Biochemistry, 50, 2011
|
|
7YZ9
 
 | | Structure of catalytic domain of Rv1625c bound to nanobody NB4 | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, Adenylate cyclase, GLYCEROL, ... | | Authors: | Khanppnavar, B, Mehta, V.J, Iype, T, Korkhov, V.M. | | Deposit date: | 2022-02-19 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of Mycobacterium tuberculosis Cya, an evolutionary ancestor of the mammalian membrane adenylyl cyclases.
Elife, 11, 2022
|
|
7ZBV
 
 | |
7Z5U
 
 | | Crystal structure of the peptidase domain of collagenase G from Clostridium histolyticum in complex with a hydroxamate-based inhibitor | | Descriptor: | (2~{R})-~{N}-[2-[4-[(2-acetamidophenoxy)methyl]-1,2,3-triazol-1-yl]ethyl]-2-(2-methylpropyl)-~{N}'-oxidanyl-propanediamide, CALCIUM ION, Collagenase ColG, ... | | Authors: | Schoenauer, E, Brandstetter, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Characterization of Synthesized and FDA-Approved Inhibitors of Clostridial and Bacillary Collagenases.
J.Med.Chem., 65, 2022
|
|
1FMC
 
 | | 7-ALPHA-HYDROXYSTEROID DEHYDROGENASE COMPLEX WITH NADH AND 7-OXO GLYCOCHENODEOXYCHOLIC ACID | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE, GLYCOCHENODEOXYCHOLIC ACID | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1996-04-26 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli.
Biochemistry, 35, 1996
|
|
2H8W
 
 | | Solution structure of ribosomal protein L11 | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Choli-Papadopoulou, T, Krueger, S, Draper, D, Wang, Y.-X. | | Deposit date: | 2006-06-08 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of Free L11 and Functional Dynamics of L11 in Free, L11-rRNA(58 nt) Binary and L11-rRNA(58 nt)-thiostrepton Ternary Complexes.
J.Mol.Biol., 367, 2007
|
|
5FP6
 
 | | Structure of cyclin-dependent kinase 2 with small-molecule ligand 3-(4,7-dichloro-1H-indol-3-yl)prop-2-yn-1-ol (AT17833) in an alternate binding site. | | Descriptor: | 3-(4,7-dichloro-1H-indol-3-yl)prop-2-yn-1-ol, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Jhoti, H, Ludlow, R.F, O'Reilly, M, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1AHH
 
 | | 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE COMPLEXED WITH NAD+ | | Descriptor: | 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1995-08-25 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli.
Biochemistry, 35, 1996
|
|
5H3O
 
 | | Structure of a eukaryotic cyclic nucleotide-gated channel | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel, SODIUM ION | | Authors: | Li, M, Zhou, X, Wang, S, Michailidis, I, Gong, Y, Su, D, Li, H, Li, X, Yang, J. | | Deposit date: | 2016-10-26 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a eukaryotic cyclic-nucleotide-gated channel.
Nature, 542, 2017
|
|
1AHI
 
 | | 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE COMPLEXED WITH NADH AND 7-OXO GLYCOCHENODEOXYCHOLIC ACID | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE, GLYCOCHENODEOXYCHOLIC ACID | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1995-08-25 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli.
Biochemistry, 35, 1996
|
|
6FQ3
 
 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains in complex with lin-29A 5'UTR 13mer RNA | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase TRIM71, RNA (5'-R(*GP*GP*AP*GP*UP*CP*CP*AP*AP*CP*UP*CP*C)-3') | | Authors: | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | Deposit date: | 2018-02-13 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
2BGK
 
 | | X-Ray structure of apo-Secoisolariciresinol Dehydrogenase | | Descriptor: | RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE | | Authors: | Youn, B, Moinuddin, S.G, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Apo-Form and Binary/Ternary Complexes of Podophyllum Secoisolariciresinol Dehydrogenase, an Enzyme Involved in Formation of Health-Protecting and Plant Defense Lignans
J.Biol.Chem., 280, 2005
|
|
5FPE
 
 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand 1H-1,2,4-triazol-3-amine (AT485) in an alternate binding site. | | Descriptor: | 3-AMINO-1,2,4-TRIAZOLE, HEAT SHOCK-RELATED 70KDA PROTEIN 2 | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-28 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FPY
 
 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 5-bromo-1-methyl-1H-indole-2-carboxylic acid (AT21457) in an alternate binding site. | | Descriptor: | 5-bromo-1-methyl-1H-indole-2-carboxylic acid, SERINE PROTEASE NS3 | | Authors: | Davies, T.G, Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6ADH
 
 | |
7LLK
 
 | |
8B9D
 
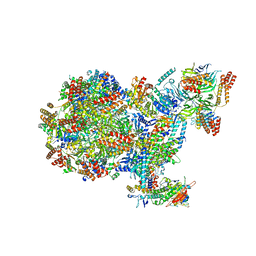 | | Human replisome bound by Pol Alpha | | Descriptor: | Cell division control protein 45 homolog, Claspin, DNA Molecule, ... | | Authors: | Jones, M.L, Yeeles, J.T.P. | | Deposit date: | 2022-10-05 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | How Pol alpha-primase is targeted to replisomes to prime eukaryotic DNA replication.
Mol.Cell, 83, 2023
|
|
7MY4
 
 | |
8B9A
 
 | |
8SYC
 
 | | Crystal structure of PDE3B in complex with GSK4394835A | | Descriptor: | MAGNESIUM ION, [3-[(4,7-dimethoxyquinolin-2-yl)carbonylamino]-5-[methyl-(phenylmethyl)carbamoyl]phenyl]-oxidanyl-oxidanylidene-boron, cGMP-inhibited 3',5'-cyclic phosphodiesterase 3B | | Authors: | Concha, N.O, Nolte, R. | | Deposit date: | 2023-05-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and SAR Study of Boronic Acid-Based Selective PDE3B Inhibitors from a Novel DNA-Encoded Library.
J.Med.Chem., 67, 2024
|
|
5A8A
 
 | | Crystal structure of the riboflavin kinase module of FAD synthetase from Corynebacterium ammoniagenes in complex with FMN and ADP (P3 2 21) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Medina, M. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Synthesis of Fmn in Prokaryotic Organisms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5A88
 
 | | Crystal structure of the riboflavin kinase module of FAD synthetase from Corynebacterium ammoniagenes in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Medina, M. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Insights Into the Synthesis of Fmn in Prokaryotic Organisms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
