1HBI
 
 | |
1MOQ
 
 | | ISOMERASE DOMAIN OF GLUCOSAMINE 6-PHOSPHATE SYNTHASE COMPLEXED WITH GLUCOSAMINE 6-PHOSPHATE | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, ... | | Authors: | Teplyakov, A. | | Deposit date: | 1997-04-11 | | Release date: | 1998-10-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Involvement of the C terminus in intramolecular nitrogen channeling in glucosamine 6-phosphate synthase: evidence from a 1.6 A crystal structure of the isomerase domain.
Structure, 6, 1998
|
|
4JWK
 
 | | Crystal structure of the complex of peptidyl-tRNA hydrolase from Acinetobacter baumannii with cytidine at 1.87 A resolution | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, Peptidyl-tRNA hydrolase | | Authors: | Kaushik, S, Singh, N, Yamini, S, Singh, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The Mode of Inhibitor Binding to Peptidyl-tRNA Hydrolase: Binding Studies and Structure Determination of Unbound and Bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii
Plos One, 8, 2013
|
|
2PCX
 
 | | Crystal structure of p53DBD(R282Q) at 1.54-angstrom Resolution | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Tu, C, Shaw, G, Ji, X. | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Impact of low-frequency hotspot mutation R282Q on the structure of p53 DNA-binding domain as revealed by crystallography at 1.54 angstroms resolution.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
260L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, PROTEIN (LYSOZYME) | | Authors: | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | Deposit date: | 1999-03-01 | | Release date: | 2000-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
4JWJ
 
 | | Crystal structure of scTrm10(84)-SAH complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2013-03-27 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate.
Nucleic Acids Res., 42, 2014
|
|
5DR1
 
 | |
4CBE
 
 | | Crystal structure of complement factors H and FHL-1 binding protein BBH06 or CRASP-2 from Borrelia burgdorferi (Native) | | Descriptor: | COMPLEMENT REGULATOR-ACQUIRING SURFACE PROTEIN 2 (CRASP-2) | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Ranka, R, Tars, K. | | Deposit date: | 2013-10-13 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Characterization of Cspz, a Complement Regulator Factor H and Fhl-1 Binding Protein from Borrelia Burgdorferi.
FEBS J., 281, 2014
|
|
3EST
 
 | | STRUCTURE OF NATIVE PORCINE PANCREATIC ELASTASE AT 1.65 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, PORCINE PANCREATIC ELASTASE, SULFATE ION | | Authors: | Meyer, E.F, Cole, G, Radhakrishnan, R, Epp, O. | | Deposit date: | 1987-09-17 | | Release date: | 1988-01-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of native porcine pancreatic elastase at 1.65 A resolutions.
Acta Crystallogr.,Sect.B, 44, 1988
|
|
7H72
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1562205518 (CHIKV_MacB-x0477) | | Descriptor: | 1-(2-hydroxyethyl)-1H-pyrazole-4-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
4J7H
 
 | | Crystal structure of EvaA, a 2,3-dehydratase in complex with dTDP-benzene and dTDP-rhamnose | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phenoxy)phosphoryl]oxy}phosphoryl]thymidine, ... | | Authors: | Holden, H.M, Kubiak, R.L, Thoden, J.B. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of EvaA: A Paradigm for Sugar 2,3-Dehydratases.
Biochemistry, 52, 2013
|
|
5MYA
 
 | | Homodimerization of Tie2 Fibronectin-like domains 1-3 in space group C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiopoietin-1 receptor, ... | | Authors: | Leppanen, V.-M, Saharinen, P, Alitalo, K. | | Deposit date: | 2017-01-26 | | Release date: | 2017-04-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of Tie2 activation and Tie2/Tie1 heterodimerization.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4C4A
 
 | | Crystal structure of mouse protein arginine methyltransferase 7 in complex with SAH | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, PROTEIN ARGININE N-METHYLTRANSFERASE 7, ... | | Authors: | Cura, V, Troffer-Charlier, N, Bonnefond, L, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-09-02 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insight Into Arginine Methylation by the Mouse Protein Arginine Methyltransferase 7: A Zinc Finger Freezes the Mimic of the Dimeric State Into a Single Active Site.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3LQA
 
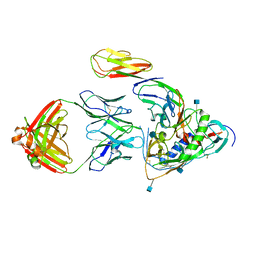 | | Crystal structure of clade C gp120 in complex with sCD4 and 21c Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, Heavy chain of anti HIV Fab from human 21c antibody, ... | | Authors: | Diskin, R, Marcovecchio, P.M, Bjorkman, P.J. | | Deposit date: | 2010-02-08 | | Release date: | 2010-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3LR2
 
 | | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Major ampullate spidroin 1 | | Authors: | Askarieh, G, Hedhammar, H, Nordling, K, Johansson, J, Knight, S.D, Rising, A, Casals, C, Saenz, A. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay.
Nature, 465, 2010
|
|
3QT4
 
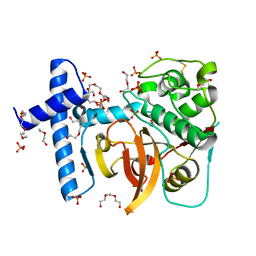 | | Structure of digestive procathepsin L 3 of Tenebrio molitor larval midgut | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Cathepsin-L-like midgut cysteine proteinase, PHOSPHATE ION, ... | | Authors: | Beton, D, Guzzo, C.R, Terra, W.R, Farah, C.S. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The 3D structure and function of digestive cathepsin L-like proteinases of Tenebrio molitor larval midgut.
Insect Biochem.Mol.Biol., 42, 2012
|
|
1HCD
 
 | | STRUCTURE OF HISACTOPHILIN IS SIMILAR TO INTERLEUKIN-1 BETA AND FIBROBLAST GROWTH FACTOR | | Descriptor: | HISACTOPHILIN | | Authors: | Habazettl, J, Gondol, D, Wiltscheck, R, Otlewski, J, Schleicher, M, Holak, T.A. | | Deposit date: | 1994-05-03 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hisactophilin is similar to interleukin-1 beta and fibroblast growth factor.
Nature, 359, 1992
|
|
2WCL
 
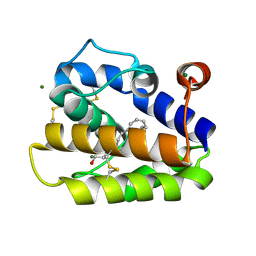 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with (8E, 10Z)-hexadecadien-1-ol | | Descriptor: | (8E,10Z)-HEXADECA-8,10-DIEN-1-OL, GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
4OZQ
 
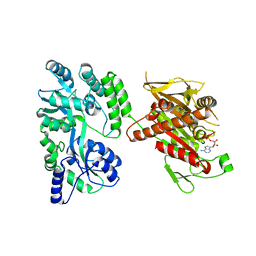 | | Crystal structure of the mouse Kif14 motor domain | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Chimera of Maltose-binding periplasmic protein and Kinesin family member 14 protein | | Authors: | Arora, K, Talje, L, Asenjo, A.B, Andersen, P, Atchia, K, Joshi, M, Sosa, H, Kwok, B.H, Allingham, J.S. | | Deposit date: | 2014-02-18 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | KIF14 binds tightly to microtubules and adopts a rigor-like conformation.
J.Mol.Biol., 426, 2014
|
|
3L5V
 
 | |
2P6X
 
 | | Crystal structure of human tyrosine phosphatase PTPN22 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 22 | | Authors: | Ugochukwu, E, Salah, E, Barr, A, Shrestha, L, Alfano, I, Burgess-Brown, N, King, O, Umeano, C, Papagrigoriou, E, Pike, A.C.W, Bunkoczi, G, Turnbull, A, Uppenberg, J, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-19 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
4JIQ
 
 | |
259L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | Descriptor: | CHLORIDE ION, COBALT (II) ION, PROTEIN (LYSOZYME) | | Authors: | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | Deposit date: | 1999-02-10 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
1ZYX
 
 | | Crystal structure of the complex of a group IIA phospholipase A2 with a synthetic anti-inflammatory agent licofelone at 1.9A resolution | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa, SULFATE ION, [6-(4-CHLOROPHENYL)-2,2-DIMETHYL-7-PHENYL-2,3-DIHYDRO-1H-PYRROLIZIN-5-YL]ACETIC ACID | | Authors: | Singh, N, Jabeen, T, Sharma, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2005-06-13 | | Release date: | 2005-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the complex of a group IIA phospholipase A2 with a synthetic anti-inflammatory agent licofelone at 1.9A resolution
To be Published
|
|
5XXO
 
 | | Crystal structure of mutant (D286N) GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorotriose | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
