3F27
 
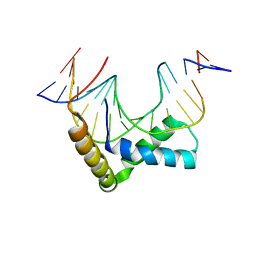 | | Structure of Sox17 Bound to DNA | | Descriptor: | DNA (5'-D(*DCP*DCP*DAP*DGP*DGP*DAP*DCP*DAP*DAP*DTP*DAP*DGP*DAP*DGP*DAP*DC)-3'), DNA (5'-D(*DGP*DTP*DCP*DTP*DCP*DTP*DAP*DTP*DTP*DGP*DTP*DCP*DCP*DTP*DGP*DG)-3'), Transcription factor SOX-17 | | Authors: | Palasingam, P, Jauch, R, Ng, C.K.L, Kolatkar, P.R. | | Deposit date: | 2008-10-29 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Structure of Sox17 Bound to DNA Reveals a Conserved Bending Topology but Selective Protein Interaction Platforms
J.Mol.Biol., 388, 2009
|
|
6LFO
 
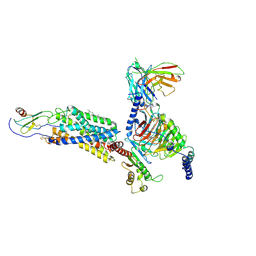 | | Cryo-EM structure of a class A GPCR monomer | | Descriptor: | C-X-C chemokine receptor type 2, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Liu, K.W, Wu, L.J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of CXC chemokine receptor 2 activation and signalling.
Nature, 585, 2020
|
|
6LFM
 
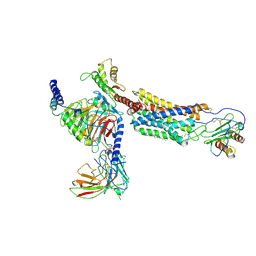 | | Cryo-EM structure of a class A GPCR | | Descriptor: | C-X-C chemokine receptor type 2, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Liu, K.W, Wu, L.J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of CXC chemokine receptor 2 activation and signalling.
Nature, 585, 2020
|
|
1CLF
 
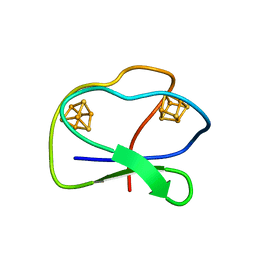 | | CLOSTRIDIUM PASTEURIANUM FERREDOXIN | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Donaire, A, Feinberg, B.A, Luchinat, C, Piccioli, M, Yuan, H. | | Deposit date: | 1995-06-21 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized 2[4Fe-4S] ferredoxin from Clostridium pasteurianum.
Eur.J.Biochem., 232, 1995
|
|
1CPR
 
 | | ST. LOUIS CYTOCHROME C' FROM THE PURPLE PHOTOTROPIC BACTERIUM, RHODOBACTER CAPSULATUS | | Descriptor: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE, ZINC ION | | Authors: | Tahirov, T.H, Misaki, S, Meyer, T.E, Cusanovich, M.A, Yasuoka, N. | | Deposit date: | 1996-04-09 | | Release date: | 1997-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of cytochrome c' from Rhodobacter capsulatus strain St Louis: an unusual molecular association induced by bridging Zn ions.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
7TAI
 
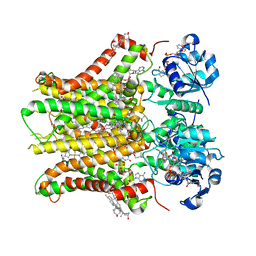 | | Structure of STEAP2 in complex with ligands | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wang, L, Chen, K.H, Zhou, M. | | Deposit date: | 2021-12-20 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of stepwise electron transfer in six-transmembrane epithelial antigen of the prostate (STEAP) 1 and 2.
Elife, 12, 2023
|
|
7FAR
 
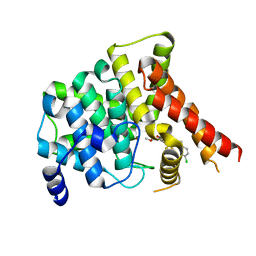 | | Crystal structure of PDE5A in complex with inhibitor L12 | | Descriptor: | 5-[bis(fluoranyl)methoxy]-2-[(4-chlorophenyl)methyl]-10-(3-methoxypropyl)-3,10-diazatricyclo[6.4.1.0^{4,13}]trideca-1,4(13),5,7-tetraen-9-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wu, D, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.40006471 Å) | | Cite: | Free energy perturbation (FEP)-guided scaffold hopping.
Acta Pharm Sin B, 12, 2022
|
|
5EYA
 
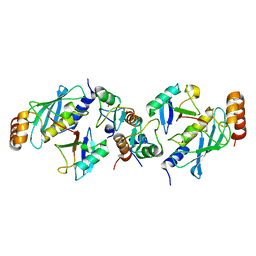 | | TRIM25 RING domain in complex with Ubc13-Ub conjugate | | Descriptor: | Polyubiquitin-B, Tripartite motif-containing 25 variant, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Pornillos, O, Sanchez, J.G. | | Deposit date: | 2015-11-24 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of TRIM25 Catalytic Activation in the Antiviral RIG-I Pathway.
Cell Rep, 16, 2016
|
|
7FAQ
 
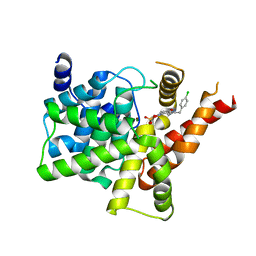 | | Crystal structure of PDE5A in complex with inhibitor L1 | | Descriptor: | 2-[(4-chlorophenyl)methyl]-5-methoxy-3,10-diazatricyclo[6.4.1.0^{4,13}]trideca-1,4(13),5,7-tetraen-9-one, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wu, D, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.200136 Å) | | Cite: | Free energy perturbation (FEP)-guided scaffold hopping.
Acta Pharm Sin B, 12, 2022
|
|
3F5O
 
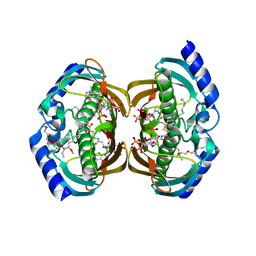 | | Crystal Structure of hTHEM2(undecan-2-one-CoA) complex | | Descriptor: | CHLORIDE ION, COENZYME A, HEXAETHYLENE GLYCOL, ... | | Authors: | Xu, H, Gong, W. | | Deposit date: | 2008-11-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mechanisms of human hotdog-fold thioesterase 2 (hTHEM2) substrate recognition and catalysis illuminated by a structure and function based analysis
Biochemistry, 48, 2009
|
|
3F7O
 
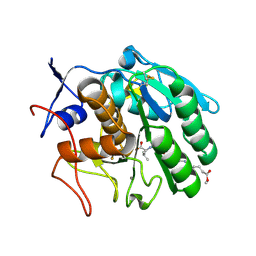 | | Crystal structure of Cuticle-Degrading Protease from Paecilomyces lilacinus (PL646) | | Descriptor: | (MSU)(ALA)(ALA)(PRO)(VAL), CALCIUM ION, Serine protease | | Authors: | Liang, L, Lou, Z, Meng, Z, Rao, Z, Zhang, K. | | Deposit date: | 2008-11-10 | | Release date: | 2009-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
8DIQ
 
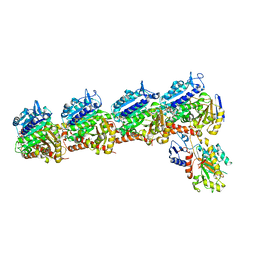 | | Tubulin-RB3_SLD-TTL in complex with SB226 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[2-(ethylamino)-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl]-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2022-06-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | SB226, an inhibitor of tubulin polymerization, inhibits paclitaxel-resistant melanoma growth and spontaneous metastasis.
Cancer Lett., 555, 2022
|
|
1YTG
 
 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | Descriptor: | HIV PROTEASE, PEPTIDE PRODUCT | | Authors: | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | Deposit date: | 1996-08-01 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
3IKG
 
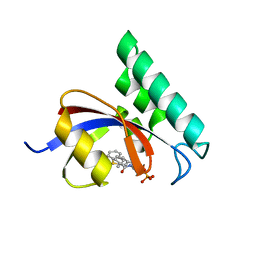 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | Descriptor: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-(3-methylphenyl)propyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Parge, H, Ferre, R.A, Greasley, S, Matthews, D. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3INH
 
 | | Bace1 with the aminohydantoin Compound R-58 | | Descriptor: | (5R)-2-amino-5-(4-fluoro-3-pyrimidin-5-ylphenyl)-3-methyl-5-[4-(trifluoromethoxy)phenyl]-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Olland, A.M, Chopra, R. | | Deposit date: | 2009-08-12 | | Release date: | 2010-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and synthesis of 5,5'-disubstituted aminohydantoins as potent and selective human beta-secretase (BACE1) inhibitors.
J.Med.Chem., 53, 2010
|
|
1D2Q
 
 | | CRYSTAL STRUCTURE OF HUMAN TRAIL | | Descriptor: | TNF-RELATED APOPTOSIS INDUCING LIGAND | | Authors: | Cha, S.-S. | | Deposit date: | 1999-09-27 | | Release date: | 2000-02-11 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 A resolution crystal structure of human TRAIL, a cytokine with selective antitumor activity.
Immunity, 11, 1999
|
|
1CEX
 
 | | STRUCTURE OF CUTINASE | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Czjzek, M, Lamzin, V, Nicolas, A, Cambillau, C. | | Deposit date: | 1997-02-18 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution (1.0 A) crystal structure of Fusarium solani cutinase: stereochemical analysis.
J.Mol.Biol., 268, 1997
|
|
6HBI
 
 | | SCAPHARCA DIMERIC HEMOGLOBIN, MUTANT T72V, DEOXY FORM | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Royer Junior, W.E. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational destabilization of the critical interface water cluster in Scapharca dimeric hemoglobin: structural basis for altered allosteric activity.
J.Mol.Biol., 284, 1998
|
|
1LH0
 
 | | Crystal Structure of Salmonella typhimurium OMP Synthase in Complex with MGPRPP and Orotate | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, MAGNESIUM ION, OMP synthase, ... | | Authors: | Fedorov, A.A, Panneerselvam, K, Shi, W, Grubmeyer, C, Almo, S.C. | | Deposit date: | 2002-04-16 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Salmonella typhimurium OMP Synthase in a Complete Substrate Complex.
Biochemistry, 51, 2012
|
|
7OJT
 
 | | Crystal structure of unliganded PatA, a membrane associated acyltransferase from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, Phosphatidylinositol mannoside acyltransferase | | Authors: | Anso, I, Wang, L, Marina, A, Paez-Perez, E.D, Perrone, S, Lowary, T.L, Trastoy, B, Guerin, M.E. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.67 Å) | | Cite: | Molecular ruler mechanism and interfacial catalysis of the integral membrane acyltransferase PatA.
Sci Adv, 7, 2021
|
|
7P76
 
 | | Re-engineered 2-deoxy-D-ribose-5-phosphate aldolase catalysing asymmetric Michael addition reactions, Schiff base complex with cinnamaldehyde | | Descriptor: | (2E)-3-phenylprop-2-enal, Deoxyribose-phosphate aldolase, GLYCEROL | | Authors: | Thunnissen, A.M.W.H, Rozeboom, H.J, Kunzendorf, A, Poelarends, G.J. | | Deposit date: | 2021-07-19 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unlocking Asymmetric Michael Additions in an Archetypical Class I Aldolase by Directed Evolution.
Acs Catalysis, 11, 2021
|
|
7P75
 
 | |
5VR6
 
 | | Structure of Human Sts-1 histidine phosphatase domain with sulfate bound | | Descriptor: | SULFATE ION, Ubiquitin-associated and SH3 domain-containing protein B | | Authors: | Zhou, W, Yin, Y, Weinheimer, A.W, Kaur, N, Carpino, N, French, J.B. | | Deposit date: | 2017-05-10 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Functional Characterization of the Histidine Phosphatase Domains of Human Sts-1 and Sts-2.
Biochemistry, 56, 2017
|
|
4Y8I
 
 | | Yeast 20S proteasome in complex with Ac-PLL-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-PLL-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4YA9
 
 | |
