4H94
 
 | | Radiation damage in lysozyme - 0.56 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7KQ6
 
 | |
4BW2
 
 | | The first bromodomain of human BRD4 in complex with 3,5 dimethylisoxaxole ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-((2-(TERT-BUTYL)PHENYL)AMINO)-7-(3,5-dimethylisoxazol-4-yl)-1,8-naphthyridine-3-carboxylic acid, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C, Mirguet, O, Lamotte, Y, Bamborough, P, Delannee, D, Bouillot, A, Gellibert, F, Krysa, G, Lewis, A, Witherington, J, Huet, P, Dudit, Y, Trottet, L, Nicodeme, E. | | Deposit date: | 2013-06-29 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Naphthyridines as Novel Bet Family Bromodomain Inhibitors.
Chemmedchem, 9, 2014
|
|
1LEN
 
 | | REFINEMENT OF TWO CRYSTAL FORMS OF LENTIL LECTIN AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, LECTIN, MANGANESE (II) ION, ... | | Authors: | Van Overberge, D, Loris, R, Wyns, L. | | Deposit date: | 1993-11-17 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of two crystal forms of lentil lectin at 1.8 A resolution.
Proteins, 20, 1994
|
|
7DDZ
 
 | | The Crystal Structure of Human Neuropeptide Y Y2 Receptor with JNJ-31020028 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Human Neuropeptide Y Y2 Receptor fusion protein, ~{N}-[4-[4-[(1~{S})-2-(diethylamino)-2-oxidanylidene-1-phenyl-ethyl]piperazin-1-yl]-3-fluoranyl-phenyl]-2-pyridin-3-yl-benzamide | | Authors: | Tang, T, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2020-10-30 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for ligand recognition of the neuropeptide Y Y 2 receptor.
Nat Commun, 12, 2021
|
|
3ZU8
 
 | | STRUCTURE OF CBM3B OF MAJOR SCAFFOLDIN SUBUNIT SCAA FROM ACETIVIBRIO CELLULOLYTICUS DETERMINED ON THE NIKEL ABSORPTION EDGE | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDIN, ... | | Authors: | Yaniv, O, Halfon, Y, Lamed, R, Frolow, F. | | Deposit date: | 2011-07-17 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of Cbm3B of the Major Scaffoldin Subunit Scaa from Acetivibrio Cellulolyticus
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5UUL
 
 | | Human Bfl-1 in complex with PUMA BH3 | | Descriptor: | Bcl-2-binding component 3, Bcl-2-related protein A1, SULFATE ION | | Authors: | Jenson, J.M, Grant, R.A, Keating, A.E. | | Deposit date: | 2017-02-17 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Epistatic mutations in PUMA BH3 drive an alternate binding mode to potently and selectively inhibit anti-apoptotic Bfl-1.
Elife, 6, 2017
|
|
6MLT
 
 | | Crystal structure of the V. cholerae biofilm matrix protein Bap1 | | Descriptor: | CALCIUM ION, CITRATE ANION, GLYCEROL, ... | | Authors: | Kaus, K, Biester, A, Chupp, E, Lu, K, Vidsudharomn, C, Olson, R. | | Deposit date: | 2018-09-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 angstrom crystal structure of the extracellular matrix protein Bap1 fromVibrio choleraeprovides insights into bacterial biofilm adhesion.
J.Biol.Chem., 294, 2019
|
|
1RZT
 
 | | Crystal structure of DNA polymerase lambda complexed with a two nucleotide gap DNA molecule | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*GP*GP*CP*AP*AP*CP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*G)-3', ... | | Authors: | Pedersen, L.C, Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Blanco, L, Kunkel, T.A. | | Deposit date: | 2003-12-29 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology.
Mol.Cell, 13, 2004
|
|
7DF7
 
 | |
7L4G
 
 | |
5UL5
 
 | | Crystal structure of RPE65 in complex with MB-004 and palmitate | | Descriptor: | (1R)-3-amino-1-{3-[(2-propylpentyl)oxy]phenyl}propan-1-ol, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, FE (II) ION, ... | | Authors: | Kiser, P.D, Palczewski, K. | | Deposit date: | 2017-01-24 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Tuning of Visual Cycle Modulator Pharmacodynamics.
J. Pharmacol. Exp. Ther., 362, 2017
|
|
1SBW
 
 | | CRYSTAL STRUCTURE OF MUNG BEAN INHIBITOR LYSINE ACTIVE FRAGMENT COMPLEX WITH BOVINE BETA-TRYPSIN AT 1.8A RESOLUTION | | Descriptor: | CALCIUM ION, PROTEIN (BETA-TRYPSIN), PROTEIN (MUNG BEAN INHIBITOR LYSIN ACTIVE FRAGMENT), ... | | Authors: | Huang, Q, Zhu, Y, Chi, C, Tang, Y. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of mung bean inhibitor lysine active fragment complex with bovine beta-trypsin at 1.8A resolution.
J.Biomol.Struct.Dyn., 16, 1999
|
|
5ULN
 
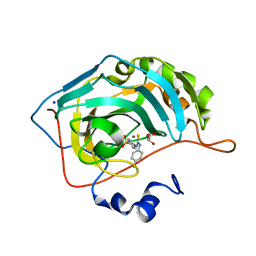 | | Synthesis of novel seleno ureido containing compounds as SLC-0111 analogs. Investigations on carbonic anhydrases activity, glutathione peroxidase and X-ray crystallography | | Descriptor: | 4-{[(4-fluorophenyl)carbamothioyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Peat, T.S, Angeli, A, Tanini, D, Bartolucci, G, Capperucci, A, Supuran, C.T, Carta, F. | | Deposit date: | 2017-01-25 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of New Selenoureido Analogues of 4-(4-Fluorophenylureido)benzenesulfonamide as Carbonic Anhydrase Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
2V5J
 
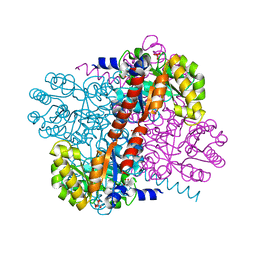 | | Apo Class II aldolase HpcH | | Descriptor: | 2,4-DIHYDROXYHEPT-2-ENE-1,7-DIOIC ACID ALDOLASE, GLYCEROL, PHOSPHATE ION | | Authors: | Rea, D, Fulop, V, Bugg, T.D.H, Roper, D.I. | | Deposit date: | 2007-07-05 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Mechanism of Hpch: A Metal Ion Dependent Class II Aldolase from the Homoprotocatechuate Degradation Pathway of Escherichia Coli.
J.Mol.Biol., 373, 2007
|
|
6SIJ
 
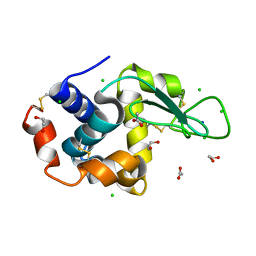 | | SAD structure of Hen Egg White Lysozyme recovered by continuous rotation data collection and multivariate analysis of Friedel pairs | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Garcia-Bonete, M.J, Katona, G. | | Deposit date: | 2019-08-10 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61018026 Å) | | Cite: | Bayesian machine learning improves single-wavelength anomalous diffraction phasing.
Acta Crystallogr.,Sect.A, 75, 2019
|
|
4H9F
 
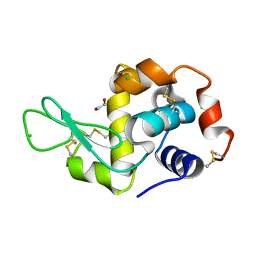 | | Radiation damage study of lysozyme - 0.91 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2003 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2VAE
 
 | | Fast maturing red fluorescent protein, DsRed.T4 | | Descriptor: | 1,2-ETHANEDIOL, RED FLUORESCENT PROTEIN | | Authors: | Strongin, D.E, Bevis, B, Khuong, N, Downing, M.E, Strack, R.L, Sundaram, K, Glick, B.S, Keenan, R.J. | | Deposit date: | 2007-08-31 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Rearrangements Near the Chromophore Influence the Maturation Speed and Brightness of Dsred Variants.
Protein Eng.Des.Sel., 20, 2007
|
|
4GLD
 
 | | Crystal Structure of the complex of type II phospholipase A2 with a designed peptide inhibitor Phe - Leu - Ala - Tyr - Lys at 1.69 A resolution | | Descriptor: | ACETATE ION, FLAYK, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Shukla, P.K, Sinha, M, Dey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-14 | | Release date: | 2012-08-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal Structure of the complex of type II phospholipase A2 with a designed peptide inhibitor Phe - Leu - Ala - Tyr - Lys at 1.69 A resolution
To be Published
|
|
1NOT
 
 | | THE 1.2 ANGSTROM STRUCTURE OF G1 ALPHA CONOTOXIN | | Descriptor: | GI ALPHA CONOTOXIN | | Authors: | Guddat, L.W, Shan, L, Martin, J.L, Edmundson, A.B, Gray, W.R. | | Deposit date: | 1996-05-02 | | Release date: | 1996-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Three-dimensional structure of the alpha-conotoxin GI at 1.2 A resolution
Biochemistry, 35, 1996
|
|
6SAF
 
 | | The Fk1 domain of FKBP51 in complex with (S)-(R)-3-(3,4-dimethoxyphenyl)-1-(3-(2-morpholinoethoxy)phenyl)propyl 1-((1R,4aR,8aR)-4-oxodecahydronaphthalene-1-carbonyl)piperidine-2-carboxylate | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, [(1~{R})-3-(3,4-dimethoxyphenyl)-1-[3-(2-morpholin-4-ylethoxy)phenyl]propyl] (2~{S})-1-[[(1~{R},4~{a}~{R},8~{a}~{R})-4-oxidanylidene-2,3,4~{a},5,6,7,8,8~{a}-octahydro-1~{H}-naphthalen-1-yl]carbonyl]piperidine-2-carboxylate | | Authors: | Feng, X, Sippel, C, Knaup, F, Bracher, A, Staibano, S, Romano, M.F, Hausch, F. | | Deposit date: | 2019-07-16 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Novel Decalin-Based Bicyclic Scaffold for FKBP51-Selective Ligands.
J.Med.Chem., 63, 2020
|
|
2BVB
 
 | | The C-terminal domain from Micronemal Protein 1 (MIC1) from Toxoplasma Gondii | | Descriptor: | MICRONEMAL PROTEIN 1 | | Authors: | Saouros, S, Edwards-Jones, B, Reiss, M, Sawmynaden, K, Cota, E, Simpson, P, Dowse, T.J, Jakle, U, Ramboarina, S, Shivarattan, T, Matthews, S, Soldati-Favre, D. | | Deposit date: | 2005-06-23 | | Release date: | 2005-10-12 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A Novel Galectin-Like Domain from Toxoplasma Gondll Micronemal Protein 1 Assists the Folding, Assembly,and Transport of a Cell-Adhesion Complex.
J.Biol.Chem., 280, 2005
|
|
7VWJ
 
 | | Matrix arm of deactive state CI from rotenone-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-10 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4DB7
 
 | | Crystal structure of B. anthracis DHPS with compound 25 | | Descriptor: | 3-(7-amino-4,5-dioxo-1,4,5,6-tetrahydropyrimido[4,5-c]pyridazin-3-yl)propanoic acid, Dihydropteroate Synthase, SULFATE ION | | Authors: | Hammoudeh, D, Lee, R.E, White, S.W. | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Novel Pyrimido[4,5-c]pyridazine Derivatives as Dihydropteroate Synthase Inhibitors with Increased Affinity.
Chemmedchem, 7, 2012
|
|
1NMI
 
 | | Solution structure of the imidazole complex of iso-1 cytochrome c | | Descriptor: | Cytochrome c, iso-1, HEME C, ... | | Authors: | Yao, Y, Tong, Y, Liu, G, Wang, J, Zheng, J, Tang, W. | | Deposit date: | 2003-01-10 | | Release date: | 2003-02-04 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the imidazole complex of iso-1 cytochrome c
To be Published
|
|
