2P63
 
 | |
2BFV
 
 | |
2A3P
 
 | | Structure of Desulfovibrio desulfuricans G20 tetraheme cytochrome with bound molybdate | | Descriptor: | COG3005: Nitrate/TMAO reductases, membrane-bound tetraheme cytochrome c subunit, HEME C, ... | | Authors: | Pattarkine, M.V, Lee, Y.-H, Tanner, J.J, Wall, J.D. | | Deposit date: | 2005-06-25 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Desulfovibrio desulfuricans G20 Tetraheme Cytochrome Structure at 1.5A and Cytochrome Interaction with Metal Complexes
J.Mol.Biol., 358, 2006
|
|
4BJC
 
 | | Crystal structure of human tankyrase 2 in complex with Rucaparib | | Descriptor: | Rucaparib, SULFATE ION, TANKYRASE-2, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-04-18 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evaluation and Structural Basis for the Inhibition of Tankyrases by Parp Inhibitors
Acs Med.Chem.Lett., 5, 2014
|
|
2GQ1
 
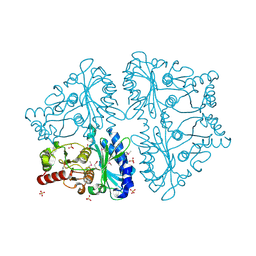 | | Crystal Structure of Recombinant Type I Fructose-1,6-bisphosphatase from Escherichia coli Complexed with Sulfate Ions | | Descriptor: | Fructose-1,6-bisphosphatase, SULFATE ION | | Authors: | Hines, J.K, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2006-04-19 | | Release date: | 2006-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Allosteric Activation Site in Escherichia coli Fructose-1,6-bisphosphatase.
J.Biol.Chem., 281, 2006
|
|
2BUH
 
 | |
5DU4
 
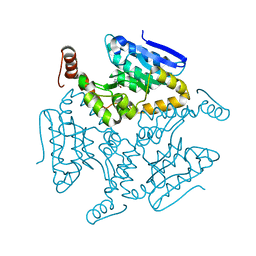 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK366A | | Descriptor: | (5R,7S)-5-(4-ethylphenyl)-N-(4-methoxybenzyl)-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
4L1M
 
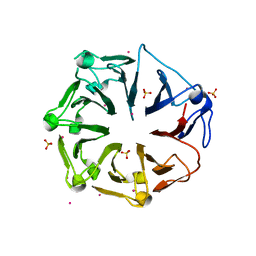 | | Structure of the first RCC1-like domain of HERC2 | | Descriptor: | E3 ubiquitin-protein ligase HERC2, SULFATE ION, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Khan, M.B, Dong, A, Hu, J, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-03 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the first RCC1-like domain of HERC2
TO BE PUBLISHED
|
|
4BPF
 
 | |
4P0B
 
 | | Crystal structure of HOIP PUB domain in complex with OTULIN PIM | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Ubiquitin thioesterase otulin | | Authors: | Akutsu, M, Schaeffer, V, Olma, M.H, Gomes, L.C, Kawasaki, M, Dikic, I. | | Deposit date: | 2014-02-20 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7005 Å) | | Cite: | Binding of OTULIN to the PUB domain of HOIP controls NF-kappa B signaling.
Mol.Cell, 54, 2014
|
|
8P64
 
 | | Co-crystal structure of PD-L1 with low molecular weight inhibitor | | Descriptor: | Programmed cell death 1 ligand 1, ~{N}-[[1-[(~{E})-2-(2-methyl-3-phenyl-phenyl)ethenyl]-1,2,3,4-tetrazol-5-yl]methyl]ethanamine | | Authors: | Plewka, J, Magiera-Mularz, K, van der Straat, R, Draijer, R, Surmiak, E, Butera, R, Land, L, Musielak, B, Domling, A. | | Deposit date: | 2023-05-25 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.312 Å) | | Cite: | 1,5-Disubstituted tetrazoles as PD-1/PD-L1 antagonists.
Rsc Med Chem, 15, 2024
|
|
6AGF
 
 | | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta1 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, X.J, li, Z.Q, Zhou, Q, Shen, H.Z, Wu, K, Huang, X.S, Chen, J.F, Zhang, J.R, Zhu, X.C, Lei, J.L, Xiong, W, Gong, H.P, Xiao, B.L, Yan, N. | | Deposit date: | 2018-08-11 | | Release date: | 2018-10-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta 1.
Science, 362, 2018
|
|
5AIE
 
 | | Not4 ring domain in complex with Ubc4 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 4, UBIQUITIN-CONJUGATING ENZYME E2 4, ZINC ION | | Authors: | Bhaskar, V, Basquin, J, Conti, E. | | Deposit date: | 2015-02-12 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Architecture of the Ubiquitylation Module of the Yeast Ccr4-not Complex.
Structure, 23, 2015
|
|
5DUF
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK729A | | Descriptor: | (5R,7S)-5-(4-ethylphenyl)-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxylic acid, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5DYZ
 
 | | Crystal structure of Asp251Gly/Gln307His mutant of cytochrome P450 BM3 in complex with N-palmitoylglycine | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, N-PALMITOYLGLYCINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Di Nardo, G, Dell'Angelo, V, Gilardi, G. | | Deposit date: | 2015-09-25 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.967 Å) | | Cite: | Subtle structural changes in the Asp251Gly/Gln307His P450 BM3 mutant responsible for new activity toward diclofenac, tolbutamide and ibuprofen.
Arch.Biochem.Biophys., 602, 2016
|
|
1EBG
 
 | |
1PDX
 
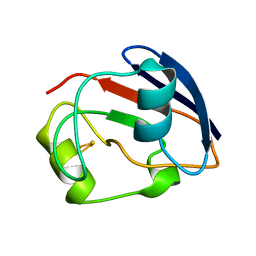 | | PUTIDAREDOXIN | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, PROTEIN (PUTIDAREDOXIN) | | Authors: | Pochapsky, T.C, Jain, N.U, Kuti, M, Lyons, T.A, Heymont, J. | | Deposit date: | 1999-02-15 | | Release date: | 1999-05-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A refined model for the solution structure of oxidized putidaredoxin.
Biochemistry, 38, 1999
|
|
2D2K
 
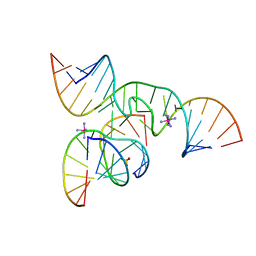 | | Crystal Structure of a minimal, native (U39) all-RNA hairpin ribozyme | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2005-09-11 | | Release date: | 2005-11-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn
Biochemistry, 44, 2005
|
|
2D3D
 
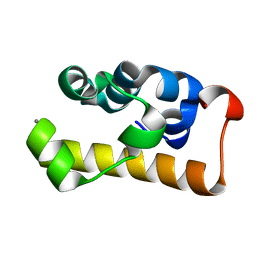 | | crystal structure of the RNA binding SAM domain of saccharomyces cerevisiae Vts1 | | Descriptor: | CALCIUM ION, Vts1 protein | | Authors: | Aviv, T, Amborski, A.N, Zhao, X.S, Kwan, J.J, Johnson, P.E, Sicheri, F, Donaldson, L.W. | | Deposit date: | 2005-09-27 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The NMR and X-ray Structures of the Saccharomyces cerevisiae Vts1 SAM Domain Define a Surface for the Recognition of RNA Hairpins
J.Mol.Biol., 356, 2006
|
|
5DU6
 
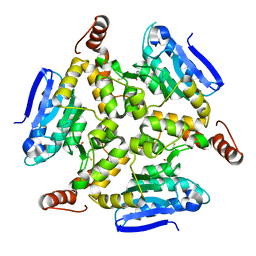 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK059A. | | Descriptor: | (5R,7R)-5-(4-ethylphenyl)-N-(4-fluorobenzyl)-7-methyl-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
4XI1
 
 | | Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, wild-type | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase LubX, GLYCEROL, ... | | Authors: | Stogios, P.J, Quaile, T, Skarina, T, Cuff, M, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-06 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.983 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
3QXU
 
 | |
2D2L
 
 | | Crystal Structure of a minimal, all-RNA hairpin ribozyme with a propyl linker (C3) at position U39 | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2005-09-11 | | Release date: | 2005-11-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn
Biochemistry, 44, 2005
|
|
1JMF
 
 | |
5B63
 
 | | Crystal structures of E.coli arginyl-tRNA synthetase (ArgRS) in complex with substrate tRNA(Arg) | | Descriptor: | Arginine--tRNA ligase, tRNA-Arg | | Authors: | Zhou, M, Ye, S, Stephen, P, Zhang, R, Wang, E.D, Giege, R, Lin, S.X. | | Deposit date: | 2016-05-24 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of E.coli arginyl-tRNA synthetase (ArgRS) in complex with substrate tRNA(Arg)
To Be Published
|
|
