5OID
 
 | | Complex Trichoplax STIL-NTD:human CEP85 coiled coil domain 4 | | Descriptor: | Centrosomal protein of 85 kDa, Putative uncharacterized protein | | Authors: | van Breugel, M. | | Deposit date: | 2017-07-18 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Direct binding of CEP85 to STIL ensures robust PLK4 activation and efficient centriole assembly.
Nat Commun, 9, 2018
|
|
6T62
 
 | |
6TCT
 
 | |
6TD1
 
 | | Crystal structure of VNRX-5133 (taniborbactam) bound to KPC-2 | | Descriptor: | (3~{R})-3-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-11-07 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Cyclic boronates as versatile scaffolds for KPC-2 beta-lactamase inhibition.
Rsc Med Chem, 11, 2020
|
|
6TD9
 
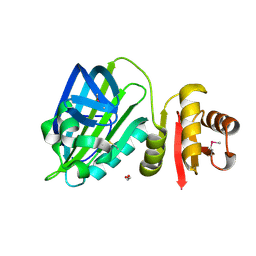 | | X-ray structure of mature PA1624 from Pseudomonas aeruginosa PAO1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PA1624, ... | | Authors: | Feiler, C.G, Blankenfeldt, W. | | Deposit date: | 2019-11-08 | | Release date: | 2019-12-04 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The hypothetical periplasmic protein PA1624 from Pseudomonas aeruginosa folds into a unique two-domain structure.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5OIQ
 
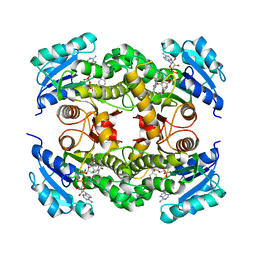 | | InhA (T2A mutant) complexed with 2,6-dimethyl-3-phenylpyridin-4(1H)-one | | Descriptor: | 2,6-dimethyl-3-phenyl-1~{H}-pyridin-4-one, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Convery, M.A. | | Deposit date: | 2017-07-19 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Screening of a Novel Fragment Library with Functional Complexity against Mycobacterium tuberculosis InhA.
ChemMedChem, 13, 2018
|
|
6T6D
 
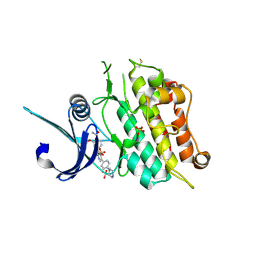 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2149 | | Descriptor: | 2-methoxy-4-[4-methyl-5-(4-piperazin-1-ylphenyl)pyridin-3-yl]benzamide, Activin receptor type I, SULFATE ION | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-18 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Targeting ALK2: An Open Science Approach to Developing Therapeutics for the Treatment of Diffuse Intrinsic Pontine Glioma.
J.Med.Chem., 63, 2020
|
|
5O2D
 
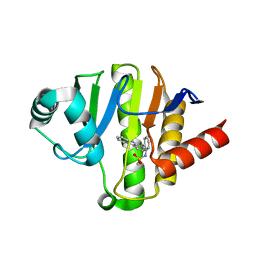 | | PARP14 Macrodomain 2 with inhibitor | | Descriptor: | Poly [ADP-ribose] polymerase 14, ~{N}-[2-(9~{H}-carbazol-1-yl)phenyl]methanesulfonamide | | Authors: | Uth, K, Schuller, M, Sieg, C, Wang, J, Krojer, T, Knapp, S, Riedels, K, Bracher, F, Edwards, A.M, Arrowsmith, C, Bountra, C, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-05-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a Selective Allosteric Inhibitor Targeting Macrodomain 2 of Polyadenosine-Diphosphate-Ribose Polymerase 14.
ACS Chem. Biol., 12, 2017
|
|
5O6G
 
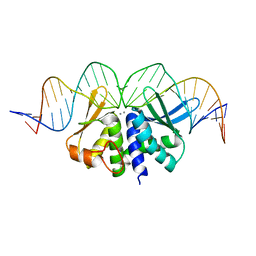 | |
5OKB
 
 | | High resolution structure of native Gan1D, a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus | | Descriptor: | GLYCEROL, IMIDAZOLE, PHOSPHATE ION, ... | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.331 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
5OL4
 
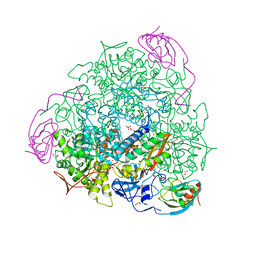 | | 1.28 A resolution of Sporosarcina pasteurii urease inhibited in the presence of NBPT | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Phosphoramidothioic O,O-acid, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2017-07-26 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Urease Inhibition in the Presence of N-(n-Butyl)thiophosphoric Triamide, a Suicide Substrate: Structure and Kinetics.
Biochemistry, 56, 2017
|
|
6T8C
 
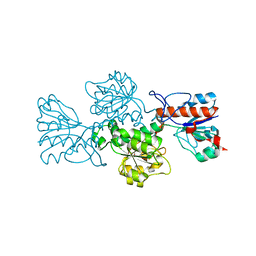 | | Crystal structure of formate dehydrogenase FDH2 enzyme from Granulicella mallensis MP5ACTX8 in the apo form. | | Descriptor: | Formate dehydrogenase | | Authors: | Robescu, M.S, Rubini, R, Filippini, F, Bergantino, B, Cendron, L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | From the Amelioration of a NADP+-dependent Formate Dehydrogenase to the Discovery of a New Enzyme: Round Trip from Theory to Practice
Chemcatchem, 2020
|
|
5OLM
 
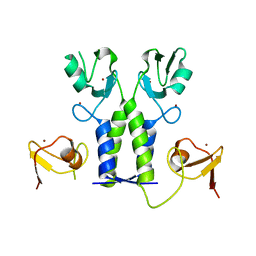 | | TRIM21 | | Descriptor: | E3 ubiquitin-protein ligase TRIM21, ZINC ION | | Authors: | James, L.C. | | Deposit date: | 2017-07-28 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Intracellular antibody signalling is regulated by phosphorylation of the Fc receptor TRIM21.
Elife, 7, 2018
|
|
6T8R
 
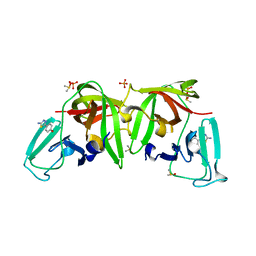 | | 3C-like protease from Southampton virus complexed with FMOPL000605a. | | Descriptor: | 4-(5-amino-1,3,4-thiadiazol-2-yl)phenol, DIMETHYL SULFOXIDE, Genome polyprotein, ... | | Authors: | Guo, J, Cooper, J.B. | | Deposit date: | 2019-10-24 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | In crystallo-screening for discovery of human norovirus 3C-like protease inhibitors.
J Struct Biol X, 4, 2020
|
|
5OLR
 
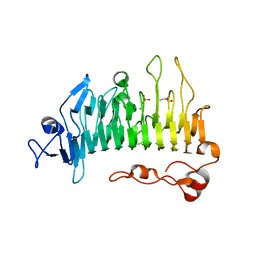 | | Rhamnogalacturonan lyase | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Rhamnogalacturonan lyase, ... | | Authors: | Basle, A, Luis, A.S, Gilbert, H.J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides.
Nat Microbiol, 3, 2018
|
|
6TAJ
 
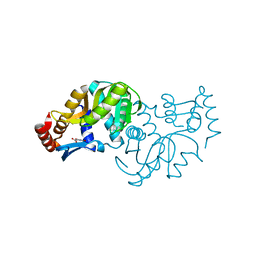 | | Crystal structure of Escherichia coli Orotate Phosphoribosyltransferase in complex with Orotic acid 1.60 Angstrom resolution | | Descriptor: | GLYCEROL, OROTIC ACID, Orotate phosphoribosyltransferase | | Authors: | Navas-Yuste, S, Lopez-Estepa, M, Gomez, S, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elucidating the Catalytic Reaction Mechanism of Orotate Phosphoribosyltransferase by Means of X-ray Crystallography and Computational Simulations
Acs Catalysis, 10, 2020
|
|
5NBG
 
 | |
5NBM
 
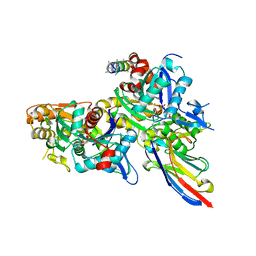 | | Crystal structure of the Arp4-N-actin(ATP-state) heterodimer bound by a nanobody | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Knoll, K.R, Eustermann, S, Hopfner, K.P. | | Deposit date: | 2017-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6TB5
 
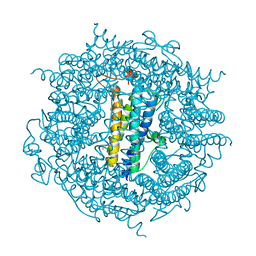 | | The crystal structure of the DPS2 from DEINOCOCCUS RADIODURANS to 1.83A resolution (sequentially soaked in CaCl2 [5mM] for 20 min, then in Ammonium iron(II) sulfate [10mM] for 2h). | | Descriptor: | CALCIUM ION, DNA protection during starvation protein 2, FE (III) ION | | Authors: | Cuypers, M.G, McSweeney, S, Romao, C.V, Mitchell, E.P. | | Deposit date: | 2019-10-31 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of the DPS2 from DEINOCOCCUS RADIODURANS to 1.83A resolution (sequentially soaked in CaCl2 [5mM] for 20 min, then in Ammonium iron(II) sulfate [10mM] for 2h).
To Be Published
|
|
6TIG
 
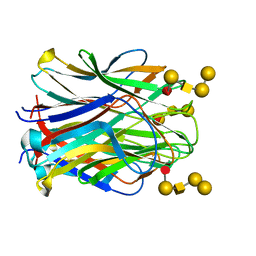 | | Structure of the N terminal domain of Bc2L-C lectin (1-131) in complex with Globo H (H-type 3) antigen | | Descriptor: | Lectin, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Varrot, A, Bermeo, R. | | Deposit date: | 2019-11-22 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | BC2L-C N-Terminal Lectin Domain Complexed with Histo Blood Group Oligosaccharides Provides New Structural Information.
Molecules, 25, 2020
|
|
5N4F
 
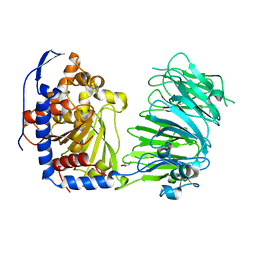 | | Prolyl oligopeptidase B from Galerina marginata - apo protein | | Descriptor: | GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
6T5M
 
 | |
5NGX
 
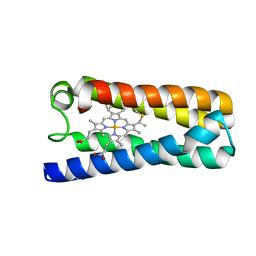 | | The 1.06 A resolution structure of the L16G mutant of ferric cytochrome c prime from Alcaligenes xylosoxidans, complexed with nitrite | | Descriptor: | Cytochrome c', GLYCEROL, HEME C, ... | | Authors: | Strange, R, Hough, M, Kekelli, D, Horrell, S, Moreno Chicano, T. | | Deposit date: | 2017-03-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Distinguishing Nitro vs Nitrito Coordination in Cytochrome c' Using Vibrational Spectroscopy and Density Functional Theory.
Inorg.Chem., 56, 2017
|
|
5N5R
 
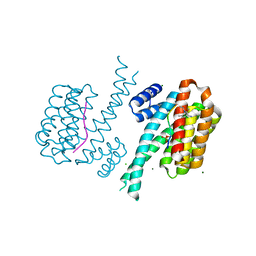 | | 14-3-3 sigma in complex with TAZ pS89 peptide and fragment NV1 | | Descriptor: | 14-3-3 protein sigma, 2-azanyl-~{N}-(2,6-dimethylphenyl)-~{N}-propan-2-yl-ethanamide, CHLORIDE ION, ... | | Authors: | Sijbesma, E, Leysen, S, Ottmann, C. | | Deposit date: | 2017-02-14 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Identification of Two Secondary Ligand Binding Sites in 14-3-3 Proteins Using Fragment Screening.
Biochemistry, 56, 2017
|
|
6T5V
 
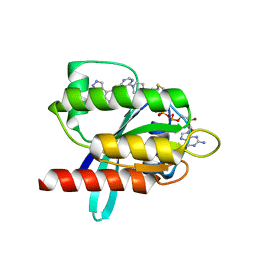 | | KRasG12C ligand complex | | Descriptor: | 1-[4-[6-chloranyl-7-(5-methyl-1~{H}-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C. | | Deposit date: | 2019-10-17 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Based Design and Pharmacokinetic Optimization of Covalent Allosteric Inhibitors of the Mutant GTPase KRASG12C.
J.Med.Chem., 63, 2020
|
|
