3DWN
 
 | | Crystal structure of the long-chain fatty acid transporter FadL mutant A77E/S100R | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Long-chain fatty acid transport protein | | Authors: | Hearn, E.M, Patel, D.R, Lepore, B.W, Indic, M, van den Berg, B. | | Deposit date: | 2008-07-22 | | Release date: | 2008-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Transmembrane passage of hydrophobic compounds through a protein channel wall.
Nature, 458, 2009
|
|
3BM6
 
 | | AmpC beta-lactamase in complex with a p.carboxyphenylboronic acid | | Descriptor: | 4-(dihydroxyboranyl)-2-({[4-(phenylsulfonyl)thiophen-2-yl]sulfonyl}amino)benzoic acid, Beta-lactamase | | Authors: | Tondi, D. | | Deposit date: | 2007-12-12 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural study of phenyl boronic acid derivatives as AmpC beta-lactamase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4GQX
 
 | | Crystal structure of EIIA(NTR) from Burkholderia pseudomallei | | Descriptor: | PTS IIA-like nitrogen-regulatory protein PtsN | | Authors: | Kim, M.-S, Shin, D.H. | | Deposit date: | 2012-08-24 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New molecular interaction of IIA(Ntr) and HPr from Burkholderia pseudomallei identified by X-ray crystallography and docking studies
Proteins, 81, 2013
|
|
3BNT
 
 | |
2R7U
 
 | | Crystal Structure of Rotavirus SA11 VP1/RNA (AAAAGCC) Complex | | Descriptor: | RNA (5'-R(*AP*A*AP*AP*GP*CP*C)-3'), RNA-dependent RNA polymerase | | Authors: | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
2R7X
 
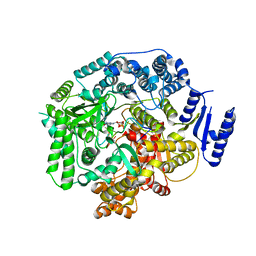 | | Crystal Structure of Rotavirus SA11 VP1/RNA (UGUGACC)/GTP complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA (5'-R(*UP*GP*UP*GP*AP*CP*C)-3'), RNA-dependent RNA polymerase | | Authors: | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
3E0M
 
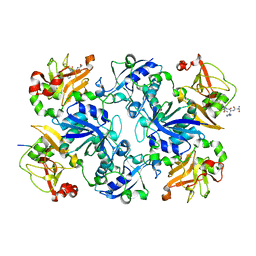 | | Crystal structure of fusion protein of MsrA and MsrB | | Descriptor: | Peptide methionine sulfoxide reductase msrA/msrB 1, Short peptide SHMAEI | | Authors: | Kim, Y.K, Hwang, K.Y. | | Deposit date: | 2008-07-31 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Kinetic Analysis of an MsrA-MsrB Fusion Protein from Streptococcus pneumoniae
Mol.Microbiol., 72, 2009
|
|
5AIU
 
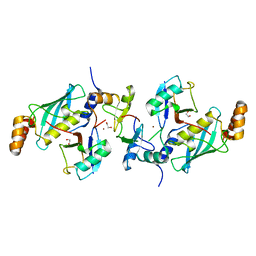 | | A complex of RNF4-RING domain, Ubc13-Ub (isopeptide crosslink) | | Descriptor: | 1,2-ETHANEDIOL, E3 UBIQUITIN-PROTEIN LIGASE RNF4, POLYUBIQUITIN-C, ... | | Authors: | Branigan, E, Naismith, J.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis for the Ring Catalyzed Synthesis of K63 Linked Ubiquitin Chains
Nat.Struct.Mol.Biol., 22, 2015
|
|
1Q2U
 
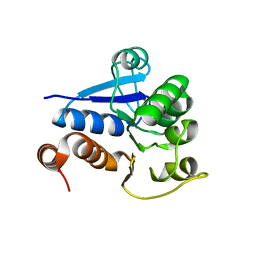 | | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Huai, Q, Sun, Y, Wang, H, Chin, L.S, Li, L, Robinson, H, Ke, H. | | Deposit date: | 2003-07-26 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease
Febs Lett., 549, 2003
|
|
7C3O
 
 | | Crystal structure of TT109 from CANDIDA ALBICANS | | Descriptor: | ACETYL COENZYME *A, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Chen, Y.P, Lei, J.H, Lu, D.R, Su, D. | | Deposit date: | 2020-05-13 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of TT109 from CANDIDA ALBICANS
To Be Published
|
|
4H6J
 
 | | Identification of Cys 255 in HIF-1 as a novel site for development of covalent inhibitors of HIF-1 /ARNT PasB domain protein-protein interaction. | | Descriptor: | ARYL HYDROCARBON NUCLEAR TRANSLOCATOR, HYPOXIA INDUCIBLE FACTOR 1-ALPHA | | Authors: | Cardoso, R, Love, R.A, Nilsson, C, Bergqvist, S, Nowlin, D, Yan, J, Liu, K, Zhu, J, Chen, P, Deng, Y.-L, Dyson, H.J, Greig, M.J, Brooun, A. | | Deposit date: | 2012-09-19 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Identification of Cys255 in HIF-1 alpha as a novel site for development of covalent inhibitors of HIF-1 alpha /ARNT PasB domain protein-protein interaction.
Protein Sci., 21, 2012
|
|
3EEG
 
 | |
4HAB
 
 | | Crystal structure of Plk1 Polo-box domain in complex with PL-49 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, PHOSPHATE ION, PL-49, ... | | Authors: | Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2012-09-26 | | Release date: | 2013-04-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Development of cyclic peptomer inhibitors targeting the polo-box domain of polo-like kinase 1.
Bioorg.Med.Chem., 21, 2013
|
|
3EHM
 
 | | Structure of BT1043 | | Descriptor: | 1,2-ETHANEDIOL, SusD homolog | | Authors: | Koropatkin, N.M, Smith, T.J. | | Deposit date: | 2008-09-13 | | Release date: | 2009-05-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a SusD homologue, BT1043, involved in mucin O-glycan utilization in a prominent human gut symbiont.
Biochemistry, 48, 2009
|
|
4GQS
 
 | | Structure of Human Microsomal Cytochrome P450 (CYP) 2C19 | | Descriptor: | (4-hydroxy-3,5-dimethylphenyl)(2-methyl-1-benzofuran-3-yl)methanone, Cytochrome P450 2C19, GLYCEROL, ... | | Authors: | Reynald, R.L, Sansen, S, Stout, C.D, Johnson, E.F. | | Deposit date: | 2012-08-23 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural Characterization of Human Cytochrome P450 2C19: ACTIVE SITE DIFFERENCES BETWEEN P450s 2C8, 2C9, AND 2C19.
J.Biol.Chem., 287, 2012
|
|
3E5F
 
 | | Crystal Structures of the SMK box (SAM-III) Riboswitch with Se-SAM | | Descriptor: | SMK box (SAM-III) Riboswitch for RNA, STRONTIUM ION, [(3S)-3-amino-4-hydroxy-4-oxo-butyl]-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl]-methyl-selanium | | Authors: | Lu, C. | | Deposit date: | 2008-08-13 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2V1N
 
 | | SOLUTION STRUCTURE OF THE REGION 51-160 OF HUMAN KIN17 REVEALS A WINGED HELIX FOLD | | Descriptor: | PROTEIN KIN HOMOLOG | | Authors: | Carlier, L, Le Maire, A, Gondry, M, Guilhaudis, L, Milazzo, I, Davoust, D, Couprie, J, Gilquin, B, Zinn-Justin, S. | | Deposit date: | 2007-05-27 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Region 51-160 of Human Kin17 Reveals an Atypical Winged Helix Domain
Protein Sci., 16, 2007
|
|
3EHN
 
 | | BT1043 with N-acetyllactosamine | | Descriptor: | SusD homolog, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Smith, T.J, Koropatkin, N.M. | | Deposit date: | 2008-09-13 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a SusD homologue, BT1043, involved in mucin O-glycan utilization in a prominent human gut symbiont.
Biochemistry, 48, 2009
|
|
364D
 
 | | 3.0 A STRUCTURE OF FRAGMENT I FROM E. COLI 5S RRNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*CP*CP*CP*AP*UP*GP*CP*GP*AP*GP*AP*GP*UP*AP*GP*G P*GP*AP*AP*CP*UP*GP*CP*CP*AP*GP*GP*CP*AP*U)-3'), RNA (5'-R(*CP*CP*GP*AP*UP*GP*GP*UP*AP*GP*UP*GP*UP*GP*GP*GP*G *UP*C)-3'), ... | | Authors: | Correll, C.C, Freeborn, B, Moore, P.B, Steitz, T.A. | | Deposit date: | 1997-12-08 | | Release date: | 1998-01-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain.
Cell(Cambridge,Mass.), 91, 1997
|
|
3ADD
 
 | | Crystal structure of O-phosphoseryl-tRNA kinase complexed with selenocysteine tRNA and AMPPNP (crystal type 3) | | Descriptor: | L-seryl-tRNA(Sec) kinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Itoh, Y, Chiba, S, Sekine, S, Yokoyama, S. | | Deposit date: | 2010-01-18 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Major Role of O-Phosphoseryl-tRNA Kinase in the UGA-Specific Encoding of Selenocysteine
Mol.Cell, 39, 2010
|
|
4E3C
 
 | | X-ray crystal structure of human IKK2 in an active conformation | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit beta | | Authors: | Polley, S, Huang, D.B, Hauenstein, A.V, Ghosh, G, Huxford, T. | | Deposit date: | 2012-03-09 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | X-ray crystal structure of human IKK2 in an active conformation
Plos.Biol., 2013
|
|
3AJH
 
 | | Crystal structure of PcyA V225D-biliverdin XIII alpha complex | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(3-ethenyl-4-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-2-ylidene]methy l]-5-[(Z)-(3-ethenyl-4-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Wada, K, Hagiwara, Y, Fukuyama, K. | | Deposit date: | 2010-06-05 | | Release date: | 2011-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | One residue substitution in PcyA leads to unexpected changes in tetrapyrrole substrate binding.
Biochem.Biophys.Res.Commun., 402, 2010
|
|
3ALW
 
 | | Crystal structure of the measles virus hemagglutinin bound to its cellular receptor SLAM (Form I, MV-H-SLAM(N102H/R108Y) fusion) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, CDw150 | | Authors: | Hashiguchi, T, Ose, T, Kubota, M, Maita, N, Kamishikiryo, J, Maenaka, K, Yanagi, Y. | | Deposit date: | 2010-08-09 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM
Nat.Struct.Mol.Biol., 18, 2011
|
|
3EKO
 
 | |
3EF1
 
 | |
