7AZP
 
 | |
7AH4
 
 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0363 | | Descriptor: | 4-chloranyl-2-(2~{H}-1,2,3-triazol-4-yl)aniline, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | Deposit date: | 2020-09-24 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Azole-Based Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AH6
 
 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0752 | | Descriptor: | 4-bromanyl-2-(4~{H}-1,2,4-triazol-3-yl)aniline, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | Deposit date: | 2020-09-24 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Azole-Based Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AH5
 
 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0706 | | Descriptor: | 4-chloranyl-2-(1~{H}-1,2,4-triazol-5-yl)aniline, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | Deposit date: | 2020-09-24 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Azole-Based Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors.
J.Med.Chem., 64, 2021
|
|
4E5Y
 
 | |
4EHI
 
 | | An X-ray Crystal Structure of a putative Bifunctional Phosphoribosylaminoimidazolecarboxamide Formyltransferase/IMP Cyclohydrolase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional purine biosynthesis protein PurH, SULFATE ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Kwok, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | An X-ray Crystal Structure of a putative Bifunctional Phosphoribosylaminoimidazolecarboxamide Formyltransferase/IMP Cyclohydrolase
TO BE PUBLISHED
|
|
6Q94
 
 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase (S156D) in complex with GDP-Man | | Descriptor: | 1,2-ETHANEDIOL, GDP-mannose 4,6 dehydratase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-17 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
2L0Y
 
 | | Complex hMia40-hCox17 | | Descriptor: | COX17 cytochrome c oxidase assembly homolog (S. cerevisiae) pseudogene (COX17), Mitochondrial intermembrane space import and assembly protein 40 | | Authors: | Bertini, I, Ciofi-Baffoni, S, Gallo, A. | | Deposit date: | 2010-07-20 | | Release date: | 2010-11-24 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6PU7
 
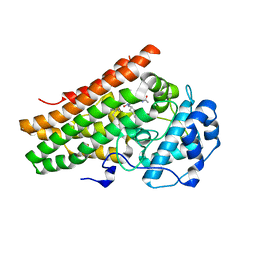 | | Human IDO1 in complex with compound 17 (N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide) | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-07-17 | | Release date: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of Amino-cyclobutarene-derived Indoleamine-2,3-dioxygenase 1 (IDO1) Inhibitors for Cancer Immunotherapy.
Acs Med.Chem.Lett., 10, 2019
|
|
2M2E
 
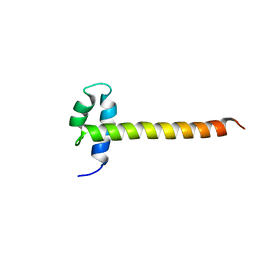 | | Solution NMR structure of the SANT domain of human DNAJC2, Northeast structural genomics consortium target HR8254a | | Descriptor: | DnaJ homolog subfamily C member 2 | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Ong, M, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-18 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the SANT domain of human DnaJC2.
To be Published
|
|
4DS3
 
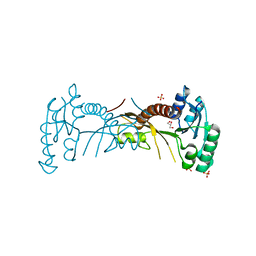 | |
4FAP
 
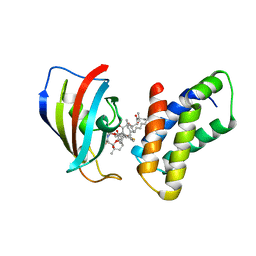 | |
2J4J
 
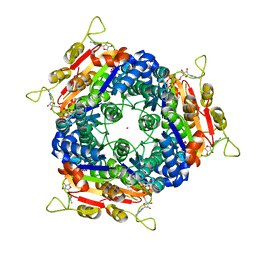 | | Crystal structure of uridylate kinase from Sulfolobus solfataricus in complex with UMP and AMPPCP to 2.1 Angstrom resolution | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, P1-(5'-ADENOSINE)P4-(5'-URIDINE)-BETA,GAMMA-METHYLENE TETRAPHOSPHATE, ... | | Authors: | Jensen, K.S, Johansson, E, Jensen, K.F. | | Deposit date: | 2006-09-01 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Enzymatic Investigation of the Sulfolobus Solfataricus Uridylate Kinase Shows Competitive Utp Inhibition and the Lack of GTP Stimulation
Biochemistry, 46, 2007
|
|
6ROP
 
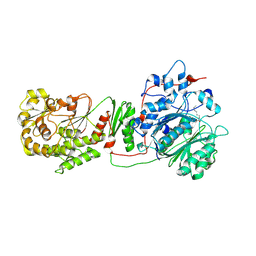 | |
2J4L
 
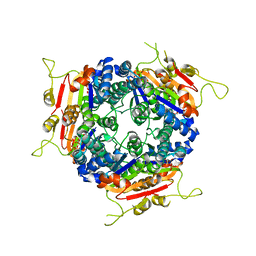 | |
4FGR
 
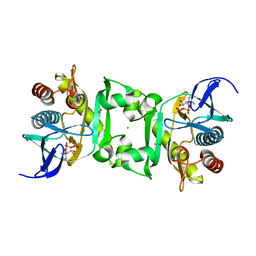 | | X-Ray Structure of SAICAR Synthetase (PurC) from Streptococcus pneumoniae complexed with ADP and Mg2+ | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Fung, L.W.-M, Johnson, M.E, Abad-Zapatero, C, Wolf, N.W. | | Deposit date: | 2012-06-04 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: |
|
|
2J4K
 
 | | Crystal structure of uridylate kinase from Sulfolobus solfataricus in complex with UMP to 2.2 Angstrom resolution | | Descriptor: | CADMIUM ION, MAGNESIUM ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Jensen, K.S, Johansson, E, Jensen, K.F. | | Deposit date: | 2006-09-01 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Enzymatic Investigation of the Sulfolobus Solfataricus Uridylate Kinase Shows Competitive Utp Inhibition and the Lack of GTP Stimulation
Biochemistry, 46, 2007
|
|
6SB2
 
 | | cryo-EM structure of mTORC1 bound to active RagA/C GTPases | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Ras-related GTP-binding protein A, ... | | Authors: | Anandapadamanaban, M, Berndt, A, Masson, G.R, Perisic, O, Williams, R.L. | | Deposit date: | 2019-07-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Architecture of human Rag GTPase heterodimers and their complex with mTORC1.
Science, 366, 2019
|
|
2JJX
 
 | | THE CRYSTAL STRUCTURE OF UMP KINASE FROM BACILLUS ANTHRACIS (BA1797) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, URIDYLATE KINASE | | Authors: | Meier, C, Carter, L.G, Mancini, E.J, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The Crystal Structure of Ump Kinase from Bacillus Anthracis (Ba1797) Reveals an Allosteric Nucleotide-Binding Site.
J.Mol.Biol., 381, 2008
|
|
2JYN
 
 | | A novel solution NMR structure of protein yst0336 from Saccharomyces cerevisiae. Northeast Structural Genomics Consortium target YT51/Ontario Centre for Structural Proteomics target yst0336 | | Descriptor: | UPF0368 protein YPL225W | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Gutmanas, A, Semest, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2007-12-14 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel solution NMR structure of protein yst0336 from Saccharomyces cerevisiae.
To be Published
|
|
6RM2
 
 | | Deoxyguanylosuccinate synthase (DgsS) structure with ATP, IMP, Magnesium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase, INOSINIC ACID, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2019-05-04 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATP, IMP, Magnesium at 2.5 Angstrom resolution
To Be Published
|
|
6SB0
 
 | | cryo-EM structure of mTORC1 bound to PRAS40-fused active RagA/C GTPases | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Proline-rich AKT1 substrate 1, ... | | Authors: | Anandapadamanaban, M, Berndt, A, Masson, G.R, Perisic, O, Williams, R.L. | | Deposit date: | 2019-07-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Architecture of human Rag GTPase heterodimers and their complex with mTORC1.
Science, 366, 2019
|
|
6R63
 
 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0358 | | Descriptor: | 4-chloranyl-2-(2~{H}-1,2,3-triazol-4-yl)phenol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Inhibition Mechanisms of Indoleamine 2,3-Dioxygenase 1 (IDO1).
J.Med.Chem., 62, 2019
|
|
2K3J
 
 | | The solution structure of human Mia40 | | Descriptor: | Mitochondrial intermembrane space import and assembly protein 40 | | Authors: | Ciofi Baffoni, S, Bertini, I, Gallo, A. | | Deposit date: | 2008-05-08 | | Release date: | 2009-02-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2NQ5
 
 | | Crystal structure of methyltransferase from Streptococcus mutans | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-30 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of methyltransferase from Streptococcus mutans
To be Published
|
|
