5HG1
 
 | | Crystal Structure of Human Hexokinase 2 with cmpd 1, a C-2-substituted glucosamine | | Descriptor: | 2-deoxy-2-{[(2E)-3-(3,4-dichlorophenyl)prop-2-enoyl]amino}-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, CITRATE ANION, ... | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5HX6
 
 | | Crystal structure of RIP1 kinase with a benzo[b][1,4]oxazepin-4-one | | Descriptor: | 5-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1,2-oxazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | DNA-Encoded Library Screening Identifies Benzo[b][1,4]oxazepin-4-ones as Highly Potent and Monoselective Receptor Interacting Protein 1 Kinase Inhibitors.
J.Med.Chem., 59, 2016
|
|
7AHI
 
 | | Substrate-engaged type 3 secretion system needle complex from Salmonella enterica typhimurium - SpaR state 2 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, Lipoprotein PrgK, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Miletic, S, Wald, J, Marlovits, T. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation
Nat Commun, 12, 2021
|
|
5HFU
 
 | | Crystal Structure of Human Hexokinase 2 with cmpd 27, a 2-amido-6-benzenesulfonamide glucosamine | | Descriptor: | Hexokinase-2, ~{N}-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-[[(4-cyanophenyl)sulfonylamino]methyl]-2,4,5-tris(oxidanyl)oxan-3-yl]-3-phenyl-benzamide | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
7AH9
 
 | | Substrate-engaged type 3 secretion system needle complex from Salmonella enterica typhimurium - SpaR state 1 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, Lipoprotein PrgK, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Miletic, S, Wald, J, Marlovits, T. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation
Nat Commun, 12, 2021
|
|
5HEX
 
 | | Crystal Structure of Human Hexokinase 2 with cmpd 30, a 2-amino-6-benzenesulfonamide glucosamine | | Descriptor: | 2-[(3-bromobenzene-1-carbonyl)amino]-6-{[(4-carboxy-5-methylfuran-2-yl)sulfonyl]amino}-2,6-dideoxy-alpha-D-glucopyranos e, Hexokinase-2 | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-06 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
7SZJ
 
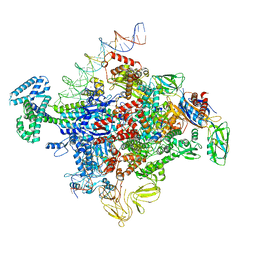 | |
6HPV
 
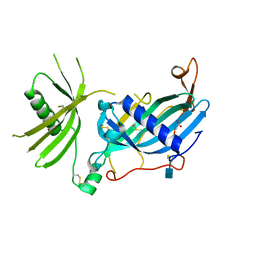 | | Crystal structure of mouse fetuin-B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Fetuin-B | | Authors: | Fahrenkamp, D, Dietzel, E, de Sanctis, D, Jovine, L. | | Deposit date: | 2018-09-22 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of mammalian plasma fetuin-B and its mechanism of selective metallopeptidase inhibition.
Iucrj, 6, 2019
|
|
8ZU2
 
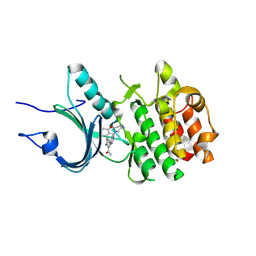 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 8g | | Descriptor: | 2-azanyl-5-[2-(1,4-diazepan-1-yl)pyridin-4-yl]-3-(2,6-dimethyl-3-oxidanyl-phenyl)benzamide, GLYCINE, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-07 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.79888582 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
8ZUL
 
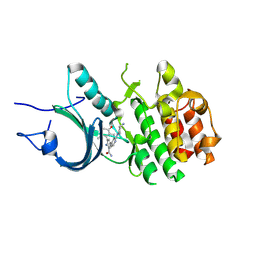 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 8m | | Descriptor: | 2-azanyl-5-[2-[(3~{R})-3-azanylpyrrolidin-1-yl]pyridin-4-yl]-3-(2,6-dimethyl-3-oxidanyl-phenyl)benzamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.80026162 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
8ZTX
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 6b | | Descriptor: | 2-azanyl-3-(2,6-dimethyl-3-oxidanyl-phenyl)-5-pyridin-4-yl-benzamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-07 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.70033228 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
8ZUD
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 8f | | Descriptor: | 2-azanyl-3-(2,6-dimethyl-3-oxidanyl-phenyl)-5-(2-morpholin-4-ylpyridin-4-yl)benzamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-08 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.50510085 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
8D9H
 
 | | gRAMP-TPR-CHAT match PFS target RNA(Craspase) | | Descriptor: | CHAT domain protein, PHOSPHATE ION, RAMP superfamily protein, ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9F
 
 | | gRAMP-TPR-CHAT (Craspase) | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
4DFY
 
 | |
8D97
 
 | | Apo gRAMP | | Descriptor: | RAMP superfamily protein, RNA (42-MER), ZINC ION | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9G
 
 | | gRAMP-TPR-CHAT Non match PFS target RNA(Craspase) | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (36-MER), ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
2LSS
 
 | | Solution structure of the R. rickettsii cold shock-like protein | | Descriptor: | Cold shock-like protein | | Authors: | Veldkamp, C.T, Peterson, F.C, Gerarden, K.P, Fuchs, A.M, Koch, J.M, Mueller, M.M. | | Deposit date: | 2012-05-04 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cold-shock-like protein from Rickettsia rickettsii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2MBF
 
 | |
2NB9
 
 | | Solution structure of ZitP zinc finger | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Campagne, S, Berge, M, Viollier, P.H, Allain, F.H.-T. | | Deposit date: | 2016-02-01 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Modularity and determinants of a (bi-)polarization control system from free-living and obligate intracellular bacteria.
Elife, 5, 2016
|
|
2L4N
 
 | |
5KBS
 
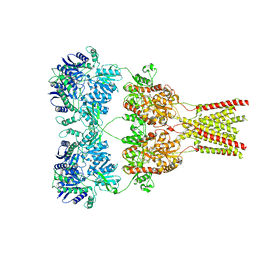 | | Cryo-EM structure of GluA2-0xSTZ at 8.7 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
5KBU
 
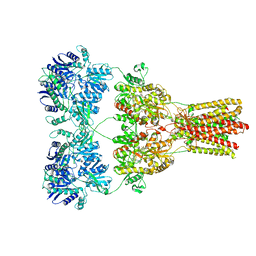 | | Cryo-EM structure of GluA2-2xSTZ complex at 7.8 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
5KBV
 
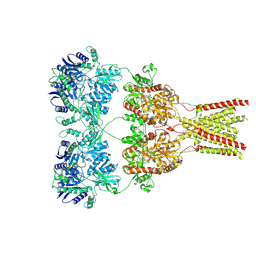 | | Cryo-EM structure of GluA2 bound to antagonist ZK200775 at 6.8 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.G, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
5KBT
 
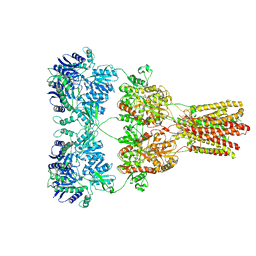 | | Cryo-EM structure of GluA2-1xSTZ complex at 6.4 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
