3HK3
 
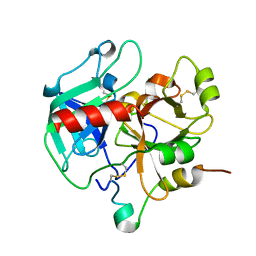 | | Crystal structure of murine thrombin mutant W215A/E217A (one molecule in the asymmetric unit) | | Descriptor: | Thrombin heavy chain, Thrombin light chain | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2009-05-22 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism of the Anticoagulant Activity of Thrombin Mutant W215A/E217A.
J.Biol.Chem., 284, 2009
|
|
1SMJ
 
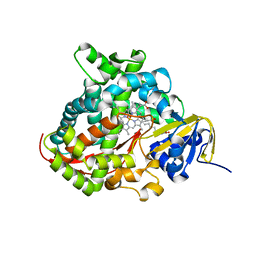 | | Structure of the A264E mutant of cytochrome P450 BM3 complexed with palmitoleate | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PALMITOLEIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Joyce, M.G, Girvan, H.M, Munro, A.W, Leys, D. | | Deposit date: | 2004-03-09 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A Single Mutation in Cytochrome P450 BM3 Induces the Conformational Rearrangement Seen upon Substrate Binding in the Wild-type Enzyme
J.Biol.Chem., 279, 2004
|
|
1MMG
 
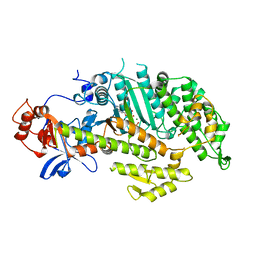 | | X-RAY STRUCTURES OF THE MGADP, MGATPGAMMAS, AND MGAMPPNP COMPLEXES OF THE DICTYOSTELIUM DISCOIDEUM MYOSIN MOTOR DOMAIN | | Descriptor: | MAGNESIUM ION, MYOSIN, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Rayment, I. | | Deposit date: | 1997-07-18 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain.
Biochemistry, 36, 1997
|
|
8P1O
 
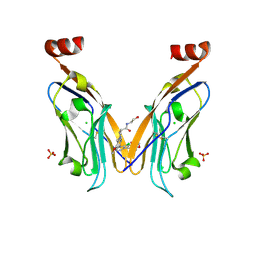 | | Solubilizer tag effect on PD-L1/inhibitor binding properties for m-terphenyl derivatives | | Descriptor: | (3~{R})-1-[[4-[2-chloranyl-3-(2,3-dihydro-1,4-benzodioxin-6-yl)phenyl]-2-methoxy-phenyl]methyl]-~{N}-(2-hydroxyethyl)pyrrolidine-3-carboxamide, CHLORIDE ION, Programmed cell death 1 ligand 1, ... | | Authors: | Plewka, J, Magiera-Mularz, K, Surmiak, E, Kalinowska-Tluscik, J, Holak, T.A. | | Deposit date: | 2023-05-12 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Solubilizer Tag Effect on PD-L1/Inhibitor Binding Properties for m -Terphenyl Derivatives.
Acs Med.Chem.Lett., 15, 2024
|
|
5LAV
 
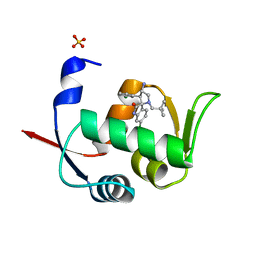 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) in complex with compound 6b | | Descriptor: | (3~{S},3'~{S},4'~{S})-4'-azanyl-6-chloranyl-3'-(3-chlorophenyl)-1'-(2,2-dimethylpropyl)spiro[1~{H}-indole-3,2'-pyrrolidine]-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
5LAY
 
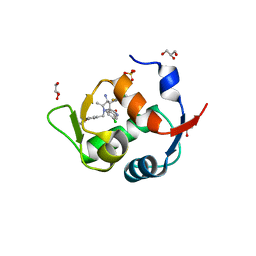 | | Discovery of New Natural-product-inspired Spiro-oxindole Compounds as Orally Active Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND 6g | | Descriptor: | (3~{S},3'~{S},4'~{S},5'~{S})-4'-azanyl-6-chloranyl-3'-(3-chloranyl-2-fluoranyl-phenyl)-1'-[(3-ethoxyphenyl)methyl]-5'-methyl-spiro[1~{H}-indole-3,2'-pyrrolidine]-2-one, E3 ubiquitin-protein ligase Mdm2, GLYCEROL, ... | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
1CQR
 
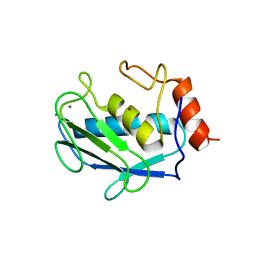 | | CRYSTAL STRUCTURE OF THE STROMELYSIN CATALYTIC DOMAIN AT 2.0 A RESOLUTION | | Descriptor: | CALCIUM ION, STROMELYSIN-1, ZINC ION | | Authors: | Chen, L, Rydel, T.J, Gu, F, Dunaway, C.M, Pikul, S, Dunham, K.M, Barnett, B.L. | | Deposit date: | 1999-08-11 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the stromelysin catalytic domain at 2.0 A resolution: inhibitor-induced conformational changes.
J.Mol.Biol., 293, 1999
|
|
5DN4
 
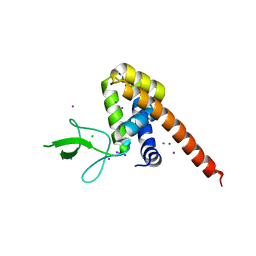 | |
1CW1
 
 | |
3DN5
 
 | | Aldose Reductase in complex with novel biarylic inhibitor | | Descriptor: | 3-[5-(3-nitrophenyl)thiophen-2-yl]propanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Klebe, G. | | Deposit date: | 2008-07-01 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Based Optimization of Aldose Reductase Inhibitors Originating from Virtual Screening
Chemmedchem, 4, 2009
|
|
4ZW7
 
 | |
4ZYQ
 
 | |
5JL4
 
 | |
5BWU
 
 | | X-RAY CRYSTAL STRUCTURE AT 2.17A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A TRIAZOLOPYRIMIDINONE COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR. | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4-bromobenzyl)amino]-5-propyl[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Branched-chain-amino-acid aminotransferase, ... | | Authors: | Somers, D.O. | | Deposit date: | 2015-06-08 | | Release date: | 2015-07-01 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The Discovery of in Vivo Active Mitochondrial Branched-Chain Aminotransferase (BCATm) Inhibitors by Hybridizing Fragment and HTS Hits.
J.Med.Chem., 58, 2015
|
|
7KSA
 
 | | Crystal structure of human CYP3A4 with the caged inhibitor | | Descriptor: | (tert-butyl {1-[(1-oxo-3-phenyl-1-{[3-(pyridin-3-yl-kappaN)prop-1-en-1-yl]amino}propan-2-yl)sulfanyl]-3-phenylpropan-2-yl}carbamate)(6,6'-dimethyl-2,2'-bipyridine-kappa~2~N~1~,N~1'~)(1~2~,2~2~:2~6~,3~2~-terpyridine-kappa~3~N~1^{1~},N~2^{1~},N~3^{1~})ruthenium, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.S. | | Deposit date: | 2020-11-21 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Photosensitive Ru(II) Complexes as Inhibitors of the Major Human Drug Metabolizing Enzyme CYP3A4.
J.Am.Chem.Soc., 143, 2021
|
|
7KS8
 
 | | Crystal structure of human CYP3A4 with the caged inhibitor | | Descriptor: | (tert-butyl {1-[(3-oxo-3-{[(pyridin-3-yl-kappaN)methyl]amino}propyl)sulfanyl]-3-phenylpropan-2-yl}carbamate)(6,6'-dimethyl-2,2'-bipyridine-kappa~2~N~1~,N~1'~)(1~2~,2~2~:2~6~,3~2~-terpyridine-kappa~3~N~1^{1~},N~2^{1~},N~3^{1~})ruthenium, Cytochrome P450 3A4, GLYCEROL, ... | | Authors: | Sevrioukova, I.S. | | Deposit date: | 2020-11-21 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Photosensitive Ru(II) Complexes as Inhibitors of the Major Human Drug Metabolizing Enzyme CYP3A4.
J.Am.Chem.Soc., 143, 2021
|
|
2YJP
 
 | | Crystal structure of the solute receptors for L-cysteine of Neisseria gonorrhoeae | | Descriptor: | 1,2-ETHANEDIOL, CYSTEINE, PUTATIVE ABC TRANSPORTER, ... | | Authors: | Bulut, H, Moniot, S, Scheffel, F, Gathmann, S, Licht, A, Saenger, W, Schneider, E. | | Deposit date: | 2011-05-23 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structures of Two Solute Receptors for L-Cystine and L-Cysteine, Respectively, of the Human Pathogen Neisseria Gonorrhoeae.
J.Mol.Biol., 415, 2012
|
|
1GAZ
 
 | | Crystal Structure of Mutant Human Lysozyme Substituted at the Surface Positions | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues at turns in the conformational stability and folding of human lysozyme.
Biochemistry, 39, 2000
|
|
4ZW8
 
 | |
1GG9
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, HIS128ASN VARIANT. | | Descriptor: | CATALASE HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-11 | | Release date: | 2000-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
4ZY0
 
 | |
1O9V
 
 | | F17-aG lectin domain from Escherichia coli in complex with a selenium carbohydrate derivative | | Descriptor: | F17-AG LECTIN DOMAIN, methyl 2-acetamido-2-deoxy-1-seleno-beta-D-glucopyranoside | | Authors: | Buts, L, De Genst, E, Loris, R, Oscarson, S, Lahmann, M, Messens, J, Brosens, E, Wyns, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2002-12-20 | | Release date: | 2003-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Fimbrial Adhesin F17-G of Enterotoxigenic Escherichia Coli Has an Immunoglobulin-Like Lectin Domain that Binds N-Acetylglucosamine
Mol.Microbiol., 49, 2003
|
|
4ZY2
 
 | | X-ray crystal structure of PfA-M17 in complex with hydroxamic acid-based inhibitor 10o | | Descriptor: | CARBONATE ION, DIMETHYL SULFOXIDE, N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluorobiphenyl-4-yl)ethyl]-2,2-dimethylpropanamide, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2015-05-21 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Potent dual inhibitors of Plasmodium falciparum M1 and M17 aminopeptidases through optimization of S1 pocket interactions.
Eur.J.Med.Chem., 110, 2016
|
|
5C4E
 
 | | Crystal structure of GTB + UDP-Glc + H-antigen acceptor | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Gagnon, S, Meloncelli, P, Zheng, R.B, Haji-Ghassemi, O, Johal, A.R, Borisova, S, Lowary, T.L, Evans, S.V. | | Deposit date: | 2015-06-18 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High Resolution Structures of the Human ABO(H) Blood Group Enzymes in Complex with Donor Analogs Reveal That the Enzymes Utilize Multiple Donor Conformations to Bind Substrates in a Stepwise Manner.
J.Biol.Chem., 290, 2015
|
|
5HKY
 
 | | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase CBL, PENTAETHYLENE GLYCOL, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form)
To be published
|
|
