6BO7
 
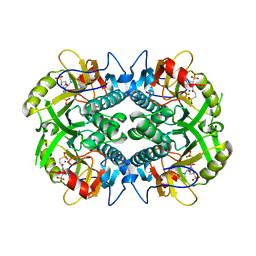 | | Crystal structure of Plasmodium vivax hypoxanthine guanine phosphoribosyltransferase in complex with [3R,4R]-4-guanin-9-yl-3-((S)-2-hydroxy-2-phosphonoethyl)oxy-1-N-(phosphonopropionyl)pyrrolidine | | Descriptor: | Hypoxanthine phosphoribosyltransferase, MAGNESIUM ION, [3-[(3~{R},4~{R})-3-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-[(2~{S})-2-oxidanyl-2-phosphono-ethoxy]pyrrolidin-1-y l]-3-oxidanylidene-propyl]phosphonic acid | | Authors: | Guddat, L.W, Keough, D.T, Rejman, D. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.856 Å) | | Cite: | Design of Plasmodium vivax Hypoxanthine-Guanine Phosphoribosyltransferase Inhibitors as Potential Antimalarial Therapeutics.
ACS Chem. Biol., 13, 2018
|
|
7XO4
 
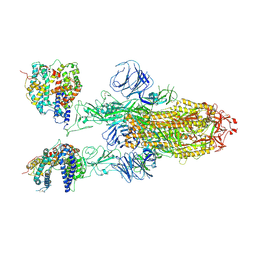 | | SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-04-30 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
1ZDX
 
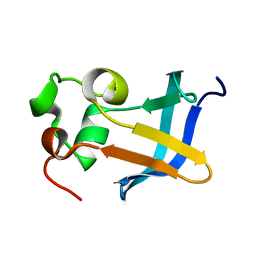 | | Solution Structure of the type 1 pilus assembly platform FimD(25-125) | | Descriptor: | Outer membrane usher protein fimD | | Authors: | Nishiyama, M, Horst, R, Herrmann, T, Vetsch, M, Bettendorff, P, Ignatov, O, Grutter, M, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
6C4J
 
 | | Ligand bound full length hUGDH with A104L substitution | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-1,2-PROPANEDIOL, ... | | Authors: | Beattie, N.R, Pioso, B.J, Wood, Z.A, Sidlo, A.M. | | Deposit date: | 2018-01-12 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Hysteresis and Allostery in Human UDP-Glucose Dehydrogenase Require a Flexible Protein Core.
Biochemistry, 57, 2018
|
|
7XOD
 
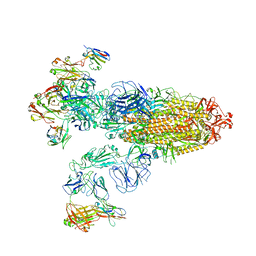 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with three JMB2002 Fab Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of JMB2002 Fab, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
6MTP
 
 | | Crystal structure of VRC42.04 Fab in complex with gp41 peptide | | Descriptor: | Antibody VRC42.04 Fab heavy chain, Antibody VRC42.04 Fab light chain, RV217 founder virus gp41 peptide | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Zhang, B, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
5UZE
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P182 | | Descriptor: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-26 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P182
To Be Published
|
|
5F2I
 
 | |
5ZLK
 
 | | Mutation in the trinuclear site of CotA-laccase: H493A mutant, PH 8.0 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase.
Chembiochem, 19, 2018
|
|
3FYG
 
 | | CRYSTAL STRUCTURE OF TETRADECA-(3-FLUOROTYROSYL)-GLUTATHIONE S-TRANSFERASE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, MU CLASS TETRADECA-(3-FLUOROTYROSYL)-GLUTATHIONE S-TRANSFERASE OF ISOENZYME | | Authors: | Xiao, G, Parsons, J.F, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1997-08-07 | | Release date: | 1999-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational changes in the crystal structure of rat glutathione transferase M1-1 with global substitution of 3-fluorotyrosine for tyrosine.
J.Mol.Biol., 281, 1998
|
|
1NVK
 
 | | T4 phage BGT in complex with UDP and a Mn2+ ion at 1.8 A resolution | | Descriptor: | DNA beta-glucosyltransferase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lariviere, L, Kurzeck, J, Gueguen-Chaignon, V, Rueger, W, Morera, S. | | Deposit date: | 2003-02-04 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the T4 phage beta-glucosyltransferase and the D100A mutant in complex with UDP-glucose: glucose binding and identification of the catalytic base for a direct displacement mechanism
J.Mol.Biol., 330, 2003
|
|
7GA2
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with POB0008 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Serine protease NS3, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.034 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7G9M
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z44584886 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Serine protease NS3, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
9I67
 
 | | StmPr1, Stenotrophomonas maltophilia Protease 1, 36 kDa alkine serine protease in complex with Chymostatin | | Descriptor: | (2~{S})-2-[[(1~{S})-1-[(6~{S})-2-azanyl-1,4,5,6-tetrahydropyrimidin-6-yl]-2-[[(2~{S})-4-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-phenyl-propan-2-yl]amino]pentan-2-yl]amino]-2-oxidanylidene-ethyl]carbamoylamino]-3-phenyl-propanoic acid, Alkaline serine protease, CALCIUM ION, ... | | Authors: | Sommer, M, Outzen, L, Negm, A, Windhorst, S, Weber, W, Betzel, C. | | Deposit date: | 2025-01-29 | | Release date: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Unveiling the structure, function and dynamics of StmPr1 in Stenotrophomonas maltophilia virulence.
Sci Rep, 15, 2025
|
|
5LLU
 
 | | Structure of the thermostabilized EAAT1 cryst-II mutant in complex with L-ASP | | Descriptor: | ASPARTIC ACID, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, SODIUM ION | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-07-28 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
5XPF
 
 | | High-resolution X-ray structure of the T26H mutant of T4 lysozyme | | Descriptor: | CHLORIDE ION, Endolysin, GLYCEROL, ... | | Authors: | Hiromoto, T, Kuroki, R. | | Deposit date: | 2017-06-01 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Neutron structure of the T26H mutant of T4 phage lysozyme provides insight into the catalytic activity of the mutant enzyme and how it differs from that of wild type.
Protein Sci., 26, 2017
|
|
4DAI
 
 | | Crystal structure of B. anthracis DHPS with compound 23 | | Descriptor: | (7-amino-4,5-dioxo-1,4,5,6-tetrahydropyrimido[4,5-c]pyridazin-3-yl)acetic acid, Dihydropteroate Synthase, SULFATE ION | | Authors: | Hammoudeh, D, Lee, R.E, White, S.W. | | Deposit date: | 2012-01-12 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Novel Pyrimido[4,5-c]pyridazine Derivatives as Dihydropteroate Synthase Inhibitors with Increased Affinity.
Chemmedchem, 7, 2012
|
|
2OIW
 
 | | The structure of a predicted thioesterase from Bacillus stearothermophilus | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, putative 4-hydroxybenzoyl-CoA thioesterase | | Authors: | Cuff, M.E, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-11 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of a predicted thioesterase from Bacillus stearothermophilus
TO BE PUBLISHED
|
|
2OKE
 
 | | High Resolution Crystal Structures of Vaccinia Virus dUTPase | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2007-01-16 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of vaccinia virus dUTPase and its nucleotide complexes.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5VRB
 
 | |
6RUU
 
 | | Pseudokinase domain of human IRAK3 | | Descriptor: | GLYCEROL, Interleukin-1 receptor-associated kinase 3, MERCURY (II) ION, ... | | Authors: | Lange, S.M, Kulathu, Y, Cohen, P. | | Deposit date: | 2019-05-29 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dimeric Structure of the Pseudokinase IRAK3 Suggests an Allosteric Mechanism for Negative Regulation.
Structure, 29, 2021
|
|
1L6W
 
 | | Fructose-6-phosphate aldolase | | Descriptor: | Fructose-6-phosphate aldolase 1, GLYCEROL | | Authors: | Thorell, S, Schuermann, M, Sprenger, G.A, Schneider, G. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of decameric fructose-6-phosphate aldolase from Escherichia coli reveals inter-subunit helix swapping as a structural basis for assembly differences in the transaldolase family.
J.Mol.Biol., 319, 2002
|
|
6VFQ
 
 | | Crystal structure of monomeric human protocadherin 10 EC1-EC4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
6VFV
 
 | | Crystal structure of human protocadherin 8 EC5-EC6 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
4HUF
 
 | | Structure of 5-chlorouracil modified A:U base pair | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*(UCL)P*TP*CP*GP*CP*G)-3'), GLYCEROL, ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs.
Nucleic Acids Res., 41, 2013
|
|
