3FHU
 
 | |
1NZU
 
 | |
3RGB
 
 | |
5KVP
 
 | |
2R37
 
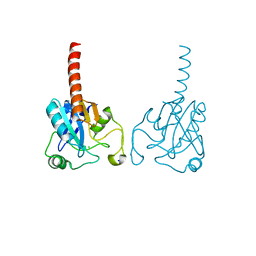 | | Crystal structure of human glutathione peroxidase 3 (selenocysteine to glycine mutant) | | Descriptor: | CHLORIDE ION, Glutathione peroxidase 3, SODIUM ION | | Authors: | Pilka, E.S, Guo, K, Gileadi, O, Rojkowa, A, von Delft, F, Pike, A.C.W, Kavanagh, K.L, Johannson, C, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-29 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human glutathione peroxidase 3 (selenocysteine to glycine mutant).
TO BE PUBLISHED
|
|
1C09
 
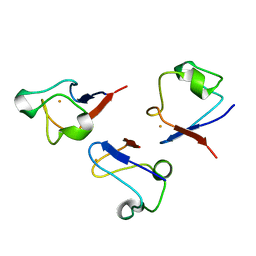 | | RUBREDOXIN V44A CP | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Min, T, Beard, B, Kang, C. | | Deposit date: | 1999-07-15 | | Release date: | 2001-02-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Modulation of the redox potential of the [Fe(SCys)(4)] site in rubredoxin by the orientation of a peptide dipole.
Biochemistry, 38, 1999
|
|
2BZG
 
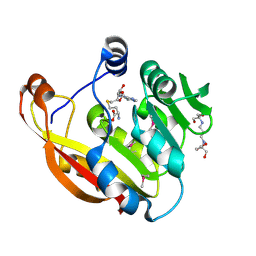 | | Crystal structure of thiopurine S-methyltransferase. | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, THIOPURINE S-METHYLTRANSFERASE | | Authors: | Battaile, K.P, Wu, H, Zeng, H, Loppnau, P, Dong, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-17 | | Release date: | 2005-08-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis of Allele Variation of Human Thiopurine-S-Methyltransferase.
Proteins, 67, 2007
|
|
2H11
 
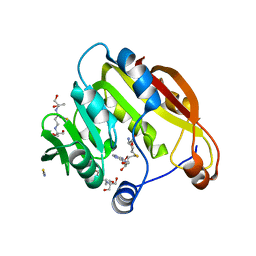 | | Amino-terminal Truncated Thiopurine S-Methyltransferase Complexed with S-Adenosyl-L-Homocysteine | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, THIOCYANATE ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2006-05-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis of allele variation of human thiopurine-S-methyltransferase.
Proteins, 67, 2007
|
|
2E0K
 
 | |
7LCU
 
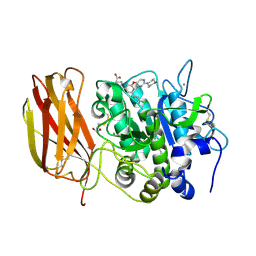 | | X-ray structure of Furin bound to BOS-318, a small molecule inhibitor | | Descriptor: | (1-{[2-(3,5-dichlorophenyl)-6-{[2-(4-methylpiperazin-1-yl)pyrimidin-5-yl]oxy}pyridin-4-yl]methyl}piperidin-4-yl)acetic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Campobasso, N, Reid, R. | | Deposit date: | 2021-01-11 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A highly selective, cell-permeable furin inhibitor BOS-318 rescues key features of cystic fibrosis airway disease.
Cell Chem Biol, 29, 2022
|
|
2QH1
 
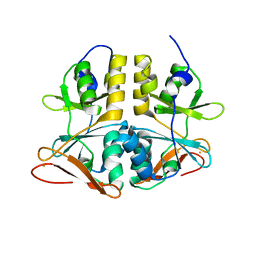 | | Structure of TA289, a CBS-rubredoxin-like protein, in its Fe+2-bound state | | Descriptor: | FE (II) ION, Hypothetical protein Ta0289 | | Authors: | Singer, A.U, Proudfoot, M, Brown, G, Xu, L, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-06-29 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of a novel family of cystathionine beta-synthase domain proteins fused to a Zn ribbon-like domain.
J.Mol.Biol., 375, 2008
|
|
4QHV
 
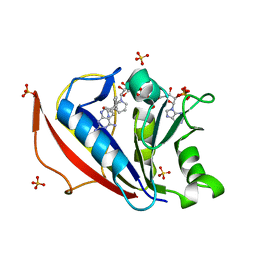 | | Crystal structure of human dihydrofolate reductase as complex with pyridopyrimidine 22 (N~6~-METHYL-N~6~-[4-(PROPAN-2-YL)PHENYL]PYRIDO[2,3-D]PYRIMIDINE-2,4,6-TRIAMINE) | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~6~-methyl-N~6~-[4-(propan-2-yl)phenyl]pyrido[2,3-d]pyrimidine-2,4,6-triamine, ... | | Authors: | Cody, V. | | Deposit date: | 2014-05-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure-activity correlations for three pyrido[2,3-d]pyrimidine antifolates binding to human and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4QJC
 
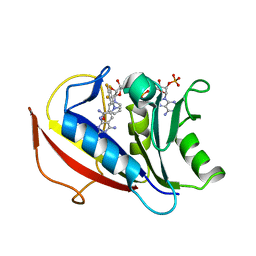 | | Human dihydrofolate reductase ternary complex with NADPH and inhibitor 26 (N~6~-METHYL-N~6~-(3,4,5-TRIFLUOROPHENYL)PYRIDO[2,3-D]PYRIMIDINE-2,4,6-TRIAMINE) | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~6~-methyl-N~6~-(3,4,5-trifluorophenyl)pyrido[2,3-d]pyrimidine-2,4,6-triamine | | Authors: | Cody, V. | | Deposit date: | 2014-06-03 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-activity correlations for three pyrido[2,3-d]pyrimidine antifolates binding to human and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
5H9K
 
 | |
1R6A
 
 | | Structure of the dengue virus 2'O methyltransferase in complex with s-adenosyl homocysteine and ribavirin 5' triphosphate | | Descriptor: | Genome polyprotein, RIBAVIRIN MONOPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Benarroch, D, Egloff, M.P, Mulard, L, Romette, J.L, Canard, B. | | Deposit date: | 2003-10-15 | | Release date: | 2004-09-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural basis for the inhibition of the NS5 dengue virus mRNA 2'-O-methyltransferase domain by ribavirin 5'-triphosphate.
J.Biol.Chem., 279, 2004
|
|
2M2S
 
 | | Solution structure of the antimicrobial peptide [Aba5,7,12,14]BTD-2 | | Descriptor: | [Aba5,7,12,14]BTD-2 | | Authors: | Conibear, A.C, Rosengren, K, Daly, N.L, Troiera Henriques, S, Craik, D.J. | | Deposit date: | 2013-01-02 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The cyclic cystine ladder in theta-defensins is important for structure and stability, but not antibacterial activity.
J.Biol.Chem., 288, 2013
|
|
4DWN
 
 | | Crystal Structure of Human BinCARD CARD | | Descriptor: | Bcl10-interacting CARD protein, SULFATE ION | | Authors: | Chen, K.-E, Kobe, B, Martin, J.L. | | Deposit date: | 2012-02-26 | | Release date: | 2013-02-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | The structure of the caspase recruitment domain of BinCARD reveals that all three cysteines can be oxidized.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5ICQ
 
 | |
2M2Y
 
 | | Solution structure of the antimicrobial peptide Btd-2[3,4] | | Descriptor: | BTD-2[3,4] | | Authors: | Conibear, A.C, Rosengren, K, Daly, N.L, Troiera Henriques, S, Craik, D.J. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | The cyclic cystine ladder in theta-defensins is important for structure and stability, but not antibacterial activity.
J.Biol.Chem., 288, 2013
|
|
1SDN
 
 | |
5H9N
 
 | | Crystal structure of LTBP1 Y114A mutant in complex with leukotriene C4 | | Descriptor: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-3-( 2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, Lipocalin AI-4 | | Authors: | Andersen, J.F. | | Deposit date: | 2015-12-28 | | Release date: | 2016-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure and Ligand-Binding Mechanism of a Cysteinyl Leukotriene-Binding Protein from a Blood-Feeding Disease Vector.
Acs Chem.Biol., 11, 2016
|
|
5H9L
 
 | | Crystal Structure of LTBP1 in complex with cleaved Leukotriene C4 | | Descriptor: | (5S,7E,9E,11Z,14Z)-5-hydroxyicosa-7,9,11,14-tetraenoic acid, GLUTATHIONE, Lipocalin AI-4 | | Authors: | Andersen, J.F. | | Deposit date: | 2015-12-28 | | Release date: | 2016-05-11 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure and Ligand-Binding Mechanism of a Cysteinyl Leukotriene-Binding Protein from a Blood-Feeding Disease Vector.
Acs Chem.Biol., 11, 2016
|
|
5HA0
 
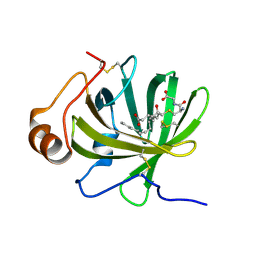 | | Crystal structure of the LTBP1 leukotriene d4 complex | | Descriptor: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-azanyl-3-(2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, Lipocalin AI-4 | | Authors: | Andersen, J.F. | | Deposit date: | 2015-12-29 | | Release date: | 2016-05-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Structure and Ligand-Binding Mechanism of a Cysteinyl Leukotriene-Binding Protein from a Blood-Feeding Disease Vector.
Acs Chem.Biol., 11, 2016
|
|
5HAE
 
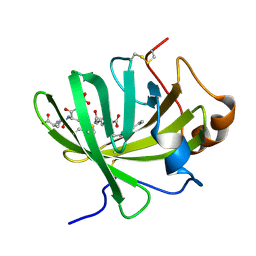 | | Crystal structure of LTBP1 LTC4 complex collected on an in-house source | | Descriptor: | (5S,7E,9E,11Z,14Z)-5-hydroxyicosa-7,9,11,14-tetraenoic acid, GLUTATHIONE, Lipocalin AI-4 | | Authors: | Andersen, J.F. | | Deposit date: | 2015-12-30 | | Release date: | 2016-05-11 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure and Ligand-Binding Mechanism of a Cysteinyl Leukotriene-Binding Protein from a Blood-Feeding Disease Vector.
Acs Chem.Biol., 11, 2016
|
|
2M1P
 
 | | [Aba5,14]BTD-2 | | Descriptor: | [Aba5,14]BTD-2 | | Authors: | Conibear, A.C, Rosengren, K, Daly, N.L, Troiera Henriques, S, Craik, D.J. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The cyclic cystine ladder in theta-defensins is important for structure and stability, but not antibacterial activity.
J.Biol.Chem., 288, 2013
|
|
