1XPP
 
 | | Crystal Structure of TA1416,DNA-directed RNA polymerase subunit L, from Thermoplasma acidophilum | | Descriptor: | ACETIC ACID, DNA-directed RNA polymerase subunit L, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of TA1416, DNA-directed RNA polymerase subunit L, from Thermoplasma acidophilum
To be Published
|
|
1XPQ
 
 | | Crystal structure of fms1, a polyamine oxidase from yeast | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1, SPERMINE | | Authors: | Huang, Q, Liu, Q, Hao, Q. | | Deposit date: | 2004-10-09 | | Release date: | 2005-04-26 | | Last modified: | 2021-12-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structures of Fms1 and its complex with spermine reveal substrate specificity.
J.Mol.Biol., 348, 2005
|
|
1XPR
 
 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic 5a-formylbicyclomycin (FB) | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', 5A-FORMYLBICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
1XPS
 
 | |
1XPT
 
 | | BOVINE RIBONUCLEASE A (PHOSPHATE-FREE) | | Descriptor: | RIBONUCLEASE A | | Authors: | Sadasivan, C, Nagendra, H.G, Vijayan, M. | | Deposit date: | 1998-02-23 | | Release date: | 1998-05-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plasticity, hydration and accessibility in ribonuclease A. The structure of a new crystal form and its low-humidity variant.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1XPU
 
 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic 5a-(3-formylphenylsulfanyl)-dihydrobicyclomycin (FPDB) | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', 5A-(3-FORMYLPHENYLSULFANYL)-DIHYDROBICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
1XPV
 
 | | Solution Structure of Northeast Structural Genomics Target Protein XcR50 from X. Campestris | | Descriptor: | hypothetical protein XCC2852 | | Authors: | Shao, Y, Acton, T.B, Liu, G, Ma, L, Shen, Y, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-09 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Northeast Structural Genomics Target Protein XcR50 from X. Campestris
To be Published
|
|
1XPW
 
 | | Solution NMR Structure of human protein HSPCO34. Northeast Structural Genomics Target HR1958 | | Descriptor: | LOC51668 protein | | Authors: | Ramelot, T.A, Xiao, R, Ma, L.C, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study.
Proteins, 75, 2009
|
|
1XPX
 
 | |
1XPY
 
 | | Structural Basis for Catalytic Racemization and Substrate Specificity of an N-Acylamino Acid Racemase Homologue from Deinococcus radiodurans | | Descriptor: | MAGNESIUM ION, N-acylamino acid racemase, N~2~-ACETYL-L-GLUTAMINE | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2004-10-10 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
1XPZ
 
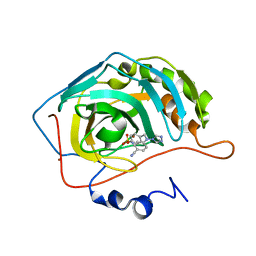 | | Structure of human carbonic anhydrase II with 4-[4-O-sulfamoylbenzyl)(4-cyanophenyl)amino]-4H-[1,2,4]-triazole | | Descriptor: | 4-{[(4-CYANOPHENYL)(4H-1,2,4-TRIAZOL-4-YL)AMINO]METHYL}PHENYL SULFAMATE, Carbonic anhydrase II, ZINC ION | | Authors: | Lloyd, M.D, Thiyagarajan, N, Ho, Y.T, Woo, L.W.L, Sutcliffe, O.B, Purohit, A, Reed, M.J, Acharya, K.R, Potter, B.V.L. | | Deposit date: | 2004-10-11 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | First Crystal Structures of Human Carbonic Anhydrase II in Complex with Dual Aromatase-Steroid Sulfatase Inhibitors(,)
Biochemistry, 44, 2005
|
|
1XQ0
 
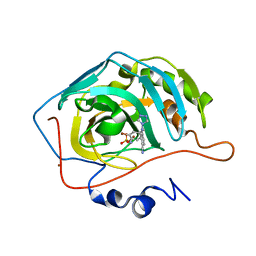 | | Structure of human carbonic anhydrase II with 4-[(3-bromo-4-O-sulfamoylbenzyl)(4-cyanophenyl)amino]-4H-[1,2,4]-triazole | | Descriptor: | 2-BROMO-4-{[(4-CYANOPHENYL)(4H-1,2,4-TRIAZOL-4-YL)AMINO]METHYL}PHENYL SULFAMATE, Carbonic anhydrase II, ZINC ION | | Authors: | Lloyd, M.D, Thiyagarajan, N, Ho, Y.T, Woo, L.W.L, Sutcliffe, O.B, Purohit, A, Reed, M.J, Acharya, K.R, Potter, B.V.L. | | Deposit date: | 2004-10-11 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | First Crystal Structures of Human Carbonic Anhydrase II in Complex with Dual Aromatase-Steroid Sulfatase Inhibitors(,)
Biochemistry, 44, 2005
|
|
1XQ1
 
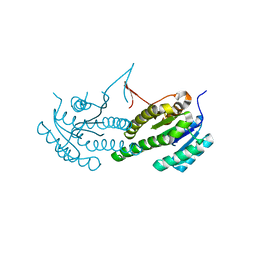 | | X-RAY STRUCTURE OF PUTATIVE TROPINONE REDUCATSE FROM ARABIDOPSIS THALIANA GENE AT1G07440 | | Descriptor: | PUTATIVE TROPINONE REDUCATSE | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-11 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-RAY STRUCTURE OF PUTATIVE TROPINONE REDUCATSE FROM ARABIDOPSIS THALIANA GENE AT1G07440
To be published
|
|
1XQ3
 
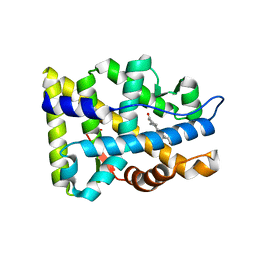 | | Crystal structure of the human androgen receptor ligand binding domain bound with R1881 | | Descriptor: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, androgen receptor | | Authors: | He, B, Gampe Jr, R.T, Kole, A.J, Hnat, A.T, Stanley, T.B, An, G, Stewart, E.L, Kalman, R.I, Minges, J.T, Wilson, E.M. | | Deposit date: | 2004-10-11 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance
Mol.Cell, 16, 2004
|
|
1XQ4
 
 | | Crystal Structure of the Putative ApaA Protein from Bordetella pertussis, Northeast Structural Genomics Target BeR40 | | Descriptor: | PHOSPHATE ION, Protein apaG | | Authors: | Forouhar, F, Yong, W, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-11 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Putative ApaA Protein from Bordetella pertussis, Northeast Structural Genomics Target BeR40
To be Published
|
|
1XQ5
 
 | |
1XQ6
 
 | | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g02240 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, unknown protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-11 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g02240
To be published
|
|
1XQ7
 
 | |
1XQ8
 
 | |
1XQ9
 
 | |
1XQA
 
 | |
1XQB
 
 | | X-Ray Structure Of YaeB from Haemophilus influenzae. Northeast Structural Genomics Research Consortium (NESGC)target IR47. | | Descriptor: | Hypothetical UPF0066 protein HI0510 | | Authors: | Benach, J, Lee, I, Forouhar, F, Kuzin, A.P, Keller, J.P, Itkin, A, Xiao, R, Acton, T, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-11 | | Release date: | 2004-10-19 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-Ray Structure Of YaeB from Haemophilus influenzae. Northeast Structural Genomics Research Consortium (NESGC) target IR47.
To be Published
|
|
1XQC
 
 | | X-ray structure of ERalpha LBD bound to a tetrahydroisoquinoline SERM ligand at 2.05A resolution | | Descriptor: | (1S)-1-{4-[(9AR)-OCTAHYDRO-2H-PYRIDO[1,2-A]PYRAZIN-2-YL]PHENYL}-2-PHENYL-1,2,3,4-TETRAHYDROISOQUINOLIN-6-OL, Estrogen receptor | | Authors: | Renaud, J, Bischoff, S.F, Buhl, T, Floersheim, P, Fournier, B, Geiser, M, Halleux, C, Kallen, J, Keller, H.J, Ramage, P. | | Deposit date: | 2004-10-12 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Selective Estrogen Receptor Modulators with Conformationally Restricted Side Chains. Synthesis and Structure-Activity Relationship of ERalpha-Selective Tetrahydroisoquinoline Ligands
J.Med.Chem., 48, 2005
|
|
1XQD
 
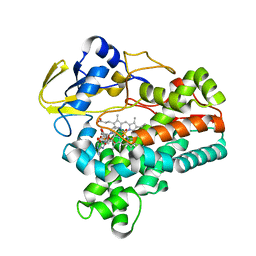 | | Crystal structure of P450NOR complexed with 3-pyridinealdehyde adenine dinucleotide | | Descriptor: | CYTOCHROME P450 55A1, NICOTINIC ACID ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oshima, R, Fushinobu, S, Takaya, N, Su, F, Wakagi, T, Shoun, H. | | Deposit date: | 2004-10-12 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for direct hydride transfer from NADH to cytochrome P450nor
J.Mol.Biol., 342, 2004
|
|
1XQE
 
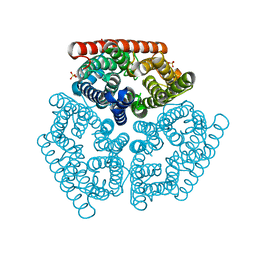 | | The mechanism of ammonia transport based on the crystal structure of AmtB of E. coli. | | Descriptor: | ACETATE ION, Probable ammonium transporter, SULFATE ION | | Authors: | Zheng, L, Kostrewa, D, Berneche, S, Winkler, F.K, Li, X.-D. | | Deposit date: | 2004-10-12 | | Release date: | 2004-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
