4POG
 
 | | MCM-ssDNA co-crystal structure | | Descriptor: | 30-mer oligo(dT), Cell division control protein 21, ZINC ION | | Authors: | Froelich, C.A, Kang, S, Epling, L.B, Bell, S.P, Enemark, E.J. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | A conserved MCM single-stranded DNA binding element is essential for replication initiation.
Elife, 3, 2014
|
|
4P6H
 
 | |
4POF
 
 | | PfMCM N-terminal domain without DNA | | Descriptor: | Cell division control protein 21, ZINC ION | | Authors: | Froelich, C.A, Kang, S, Epling, L.B, Bell, S.P, Enemark, E.J. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | A conserved MCM single-stranded DNA binding element is essential for replication initiation.
Elife, 3, 2014
|
|
4P6Z
 
 | |
4PPP
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Fluoro-Resveratrol | | Descriptor: | 5-[(E)-2-(3-fluoro-4-hydroxyphenyl)ethenyl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Parent, A.A, Hughes, T.S, Pollock, J.A, Gjyshi, O, Cavett, V, Nowak, J, Garcia-Ordonez, R.D, Houtman, R, Griffin, P.R, Kojetin, D.J, Katzenellenbogen, J.A, Conkright, M.D, Nettles, K.W. | | Deposit date: | 2014-02-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network.
Elife, 3, 2014
|
|
4PXT
 
 | |
4B9H
 
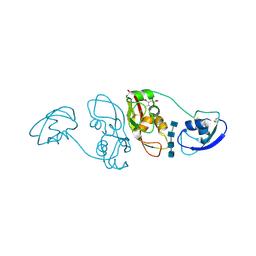 | | Cladosporium fulvum LysM effector Ecp6 in complex with a beta-1,4- linked N-acetyl-D-glucosamine tetramer: I3C heavy atom derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Saleem-Batcha, R, Sanchez-Vallet, A, Hansen, G, Thomma, B.P.H.J, Mesters, J.R. | | Deposit date: | 2012-09-04 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fungal Effector Ecp6 Outcompetes Host Immune Receptor for Chitin Binding Through Intrachain Lysm Dimerization
Elife, 2, 2013
|
|
4DQU
 
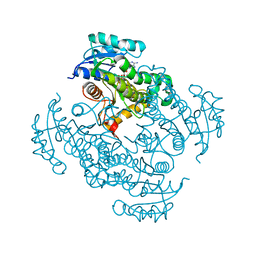 | | Mycobacterium tuberculosis InhA-D148G mutant in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pojer, F, Hartkoorn, R.C, Boy, S, Cole, S.T. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Towards a new tuberculosis drug: pyridomycin - nature's isoniazid.
EMBO Mol Med, 4, 2012
|
|
3VST
 
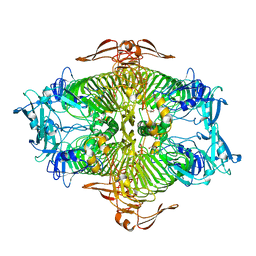 | | The complex structure of XylC with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Xylosidase | | Authors: | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | Deposit date: | 2012-05-09 | | Release date: | 2013-02-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
3VSU
 
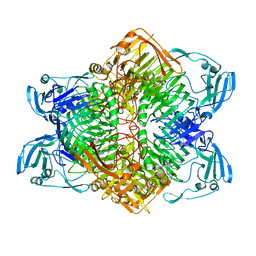 | | The complex structure of XylC with xylobiose | | Descriptor: | Xylosidase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | Deposit date: | 2012-05-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
4ALZ
 
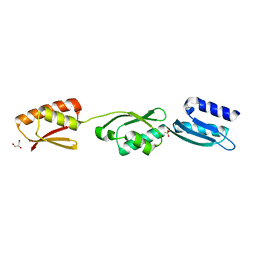 | | The Yersinia T3SS basal body component YscD reveals a different structural periplasmatic domain organization to known homologue PrgH | | Descriptor: | GLYCEROL, PHOSPHATE ION, YOP PROTEINS TRANSLOCATION PROTEIN D | | Authors: | Schmelz, S, Wisand, U, Stenta, M, Muenich, S, Widow, U, Cornelis, G.R, Heinz, D.W. | | Deposit date: | 2012-03-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | In Situ Structural Analysis of the Yersinia Enterocolitica Injectisome.
Elife, 2, 2013
|
|
4FVM
 
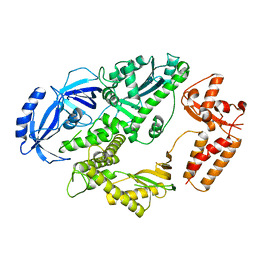 | |
4B8V
 
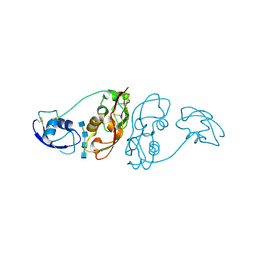 | | Cladosporium fulvum LysM effector Ecp6 in complex with a beta-1,4- linked N-acetyl-D-glucosamine tetramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Saleem-Batcha, R, Sanchez-Vallet, A, Hansen, G, Thomma, B.P.H.J, Mesters, J.R. | | Deposit date: | 2012-08-30 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Fungal Effector Ecp6 Outcompetes Host Immune Receptor for Chitin Binding Through Intrachain Lysm Dimerization
Elife, 2, 2013
|
|
4FXD
 
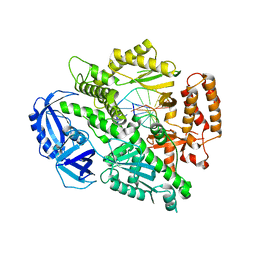 | | Crystal structure of yeast DNA polymerase alpha bound to DNA/RNA | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*CP*GP*CP*TP*GP*CP*CP*CP*GP*CP*CP*T)-3'), DNA polymerase alpha catalytic subunit A, RNA (5'-R(*AP*GP*GP*CP*GP*GP*GP*CP*AP*G)-3') | | Authors: | Perera, R.L, Torella, R, Klinge, S, Kilkenny, M.L, Maman, J.D, Pellegrini, L. | | Deposit date: | 2012-07-03 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism for priming DNA synthesis by yeast DNA Polymerase alpha
eLife, 2, 2013
|
|
4B08
 
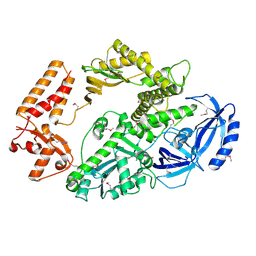 | | Yeast DNA polymerase alpha, Selenomethionine protein | | Descriptor: | DNA POLYMERASE ALPHA CATALYTIC SUBUNIT A | | Authors: | Perera, R.L, Torella, R, Klinge, S, Kilkenny, M.L, Maman, J.D, Pellegrini, L. | | Deposit date: | 2012-06-29 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Mechanism for Priming DNA Synthesis by Yeast DNA Polymerase Alpha
Elife, 2, 2013
|
|
4BL0
 
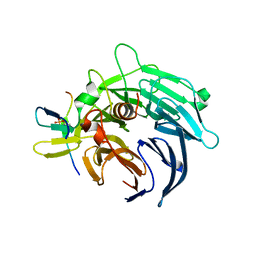 | | Crystal structure of yeast Bub3-Bub1 bound to phospho-Spc105 | | Descriptor: | CELL CYCLE ARREST PROTEIN BUB3, CHECKPOINT SERINE/THREONINE-PROTEIN KINASE BUB1, MAGNESIUM ION, ... | | Authors: | Primorac, I, Weir, J.R, Musacchio, A. | | Deposit date: | 2013-04-30 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bub3 Reads Phosphorylated Melt Repeats to Promote Spindle Assembly Checkpoint Signaling
Elife, 2, 2013
|
|
4BZI
 
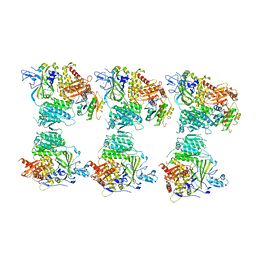 | | The structure of the COPII coat assembled on membranes | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, SAR1P, ... | | Authors: | Zanetti, G, Prinz, S, Daum, S, Meister, A, Schekman, R, Bacia, K, Briggs, J.A.G. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | The Structure of the Copii Transport-Vesicle Coat Assembled on Membranes
Elife, 2, 2013
|
|
4DTI
 
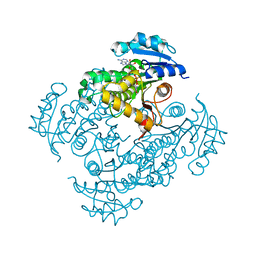 | | Mycobacterium tuberculosis InhA-S94A mutant in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pojer, F, Hartkoorn, R.C, Boy, S, Cole, S.T. | | Deposit date: | 2012-02-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards a new tuberculosis drug: pyridomycin - nature's isoniazid.
EMBO Mol Med, 4, 2012
|
|
4DRE
 
 | | Mycobacterium tuberculosis InhA in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pojer, F, Hartkoorn, R.C, Boy, S, Cole, S.T. | | Deposit date: | 2012-02-17 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Towards a new tuberculosis drug: pyridomycin - nature's isoniazid.
EMBO Mol Med, 4, 2012
|
|
4C7A
 
 | | Crystal structure of the Smoothened CRD, selenomethionine-labeled | | Descriptor: | SMOOTHENED, SODIUM ION, ZINC ION | | Authors: | Nachtergaele, S, Whalen, D.M, Mydock, L.K, Zhao, Z, Malinauskas, T, Krishnan, K, Ingham, P.W, Covey, D.F, Rohatgi, R, Siebold, C. | | Deposit date: | 2013-09-20 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of the Smoothened Extracellular Domain in Vertebrate Hedgehog Signaling
Elife, 2, 2013
|
|
3VSV
 
 | | The complex structure of XylC with xylose | | Descriptor: | Xylosidase, alpha-D-xylopyranose, beta-D-xylopyranose | | Authors: | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | Deposit date: | 2012-05-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
4BXQ
 
 | |
4BXP
 
 | |
4BZJ
 
 | | The structure of the COPII coat assembled on membranes | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Zanetti, G, Prinz, S, Daum, S, Meister, A, Schekman, R, Bacia, K, Briggs, J.A.G. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (40 Å) | | Cite: | The Structure of the Copii Transport-Vesicle Coat Assembled on Membranes
Elife, 2, 2013
|
|
4C3R
 
 | | Structure of dephosphorylated Aurora A (122-403) bound to AMPPCP | | Descriptor: | AURORA KINASE A, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Zorba, A, Kutter, S, Cho, Y.-J, Kern, D. | | Deposit date: | 2013-08-26 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Molecular Mechanism of Aurora a Kinase Autophosphorylation and its Allosteric Activation by Tpx2.
Elife, 3, 2014
|
|
