6QYS
 
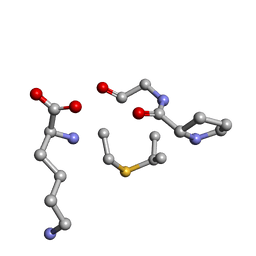 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Nisin Ring B | | Descriptor: | DBB-PRO-GLY-CYS-LYS | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
9FLC
 
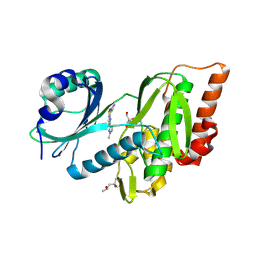 | | Crystal structure of haspin (GSG2) in complex with MU1668 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1-methylpyrazol-3-yl)-3-pyridin-4-yl-thieno[3,2-b]pyridine, GLYCEROL, ... | | Authors: | Chaikuad, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-04 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Thieno[3,2-b]pyridine: Attractive scaffold for highly selective inhibitors of underexplored protein kinases with variable binding mode.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9FLT
 
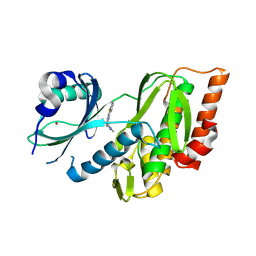 | | Crystal structure of human Haspin (GSG2) kinase bound to chemical probe MU1920 | | Descriptor: | NICKEL (II) ION, Serine/threonine-protein kinase haspin, ~{N}-(1,4-dimethylpyrazol-3-yl)-3-pyridin-4-yl-thieno[3,2-b]pyridin-5-amine | | Authors: | Kraemer, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-05 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thieno[3,2-b]pyridine: Attractive scaffold for highly selective inhibitors of underexplored protein kinases with variable binding mode.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9FLB
 
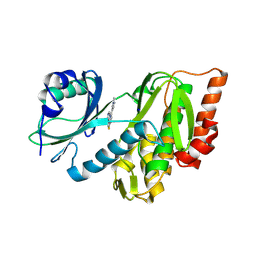 | | Crystal structure of haspin (GSG2) in complex with MU1464 | | Descriptor: | 5-(1-methylpyrazol-4-yl)-3-pyridin-4-yl-thieno[3,2-b]pyridine, SODIUM ION, Serine/threonine-protein kinase haspin | | Authors: | Chaikuad, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-04 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Thieno[3,2-b]pyridine: Attractive scaffold for highly selective inhibitors of underexplored protein kinases with variable binding mode.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9FLO
 
 | | Crystal structure of human Haspin (GSG2) kinase bound to MU2181 | | Descriptor: | NICKEL (II) ION, Serine/threonine-protein kinase haspin, ~{N}-(1,4-dimethylpyrazol-3-yl)-3-(1,2-thiazol-5-yl)thieno[3,2-b]pyridin-5-amine | | Authors: | Kraemer, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-05 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Thieno[3,2-b]pyridine: Attractive scaffold for highly selective inhibitors of underexplored protein kinases with variable binding mode.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
4XCB
 
 | | Crystal Structure of HygX from Streptomyces hygroscopicus with nickel, 2-oxoglutarate, and hygromycin B bound | | Descriptor: | 2-OXOGLUTARIC ACID, HYGROMYCIN B VARIANT, NICKEL (II) ION, ... | | Authors: | McCulloch, K.M, McCranie, E.K, Sarwar, M, Mathieu, J.L, Gitschlag, B.L, Du, Y, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2014-12-17 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6QYR
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring B, minor conformer | | Descriptor: | DAL-LEU-GLY-CYS-THR | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
3DZL
 
 | | Crystal structure of PhzA/B from Burkholderia cepacia R18194 in complex with (R)-3-oxocyclohexanecarboxylic acid | | Descriptor: | (1R)-3-oxocyclohexanecarboxylic acid, Phenazine biosynthesis protein A/B | | Authors: | Ahuja, E.G, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2008-07-30 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PhzA/B Catalyzes the Formation of the Tricycle in Phenazine Biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
9I4F
 
 | | Blood Type B-converting alpha-1,3-galactosidase PpaGal from Pedobacter panaciterrae in its apo form | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,3-galactosidase PpaGal | | Authors: | Schmoeker, O, Moeller, C, Terholsen, H, Girbardt, B, Palm, G.J, Hoppen, J, Lammers, M, Bornscheuer, U.T. | | Deposit date: | 2025-01-24 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification and Protein Engineering of Galactosidases for the Conversion of Blood Type B to Blood Type O.
Chembiochem, 26, 2025
|
|
8KHU
 
 | | Hepatitis B virus core protein Y132A mutant in complex with THPP derivatives 48 | | Descriptor: | (6~{S},7~{R})-6,7-dimethyl-3-(2-oxidanylidenepyrrolidin-1-yl)-~{N}-[3,4,5-tris(fluoranyl)phenyl]-6,7-dihydro-4~{H}-pyrazolo[1,5-a]pyrazine-5-carboxamide, Capsid protein, GLYCEROL, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2023-08-22 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4,5,6,7-Tetrahydropyrazolo[1.5-a]pyrizine Derivatives as Core Protein Allosteric Modulators (CpAMs) for the Inhibition of Hepatitis B Virus.
J.Med.Chem., 66, 2023
|
|
5AAE
 
 | | Aurora A kinase bound to an imidazopyridine inhibitor (14d) | | Descriptor: | 3-((4-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)-1H-pyrazol-1-yl)methyl)-5-methylisoxazole, AURORA KINASE A | | Authors: | McIntyre, P.J, Bayliss, R. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | 7-(Pyrazol-4-Yl)-3H-Imidazo[4,5-B]Pyridine-Based Derivatives for Kinase Inhibition: Co-Crystallisation Studies with Aurora-A Reveal Distinct Differences in the Orientation of the Pyrazole N1-Substituent.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8TN9
 
 | | Structural architecture of the acidic region of the B domain of coagulation factor V | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor V | | Authors: | Mohammed, B.M, Basore, K, Summers, B, Pelc, L.A, Di Cera, E. | | Deposit date: | 2023-08-01 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural architecture of the acidic region of the B domain of coagulation factor V.
J.Thromb.Haemost., 22, 2024
|
|
5AAD
 
 | | Aurora A kinase bound to an imidazopyridine inhibitor (7a) | | Descriptor: | 7-(1-benzyl-1H-pyrazol-4-yl)-6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridine, AURORA KINASE A, SULFATE ION | | Authors: | McIntyre, P.J, Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | 7-(Pyrazol-4-Yl)-3H-Imidazo[4,5-B]Pyridine-Based Derivatives for Kinase Inhibition: Co-Crystallisation Studies with Aurora-A Reveal Distinct Differences in the Orientation of the Pyrazole N1-Substituent.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3EXV
 
 | |
1AQ7
 
 | |
3LQK
 
 | | Crystal structure of dipicolinate synthase subunit B from Bacillus halodurans C | | Descriptor: | Dipicolinate synthase subunit B, PHOSPHATE ION | | Authors: | Nocek, B, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-09 | | Release date: | 2010-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dipicolinate synthase subunit B from Bacillus halodurans C
To be Published
|
|
3EX9
 
 | |
5AAF
 
 | | Aurora A kinase bound to an imidazopyridine inhibitor (14a) | | Descriptor: | 3-((4-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)-1H-pyrazol-1-yl)methyl)-N,N-dimethylbenzamide, AURORA KINASE A | | Authors: | McIntyre, P.J, Bayliss, R. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | 7-(Pyrazol-4-Yl)-3H-Imidazo[4,5-B]Pyridine-Based Derivatives for Kinase Inhibition: Co-Crystallisation Studies with Aurora-A Reveal Distinct Differences in the Orientation of the Pyrazole N1-Substituent.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4OUK
 
 | | Crystal structure of a C6-C4 SN3 inhibited ESTERASE B from LACTOBACILLUS RHAMNOSIS | | Descriptor: | (2R)-2,3-bis(hexyloxy)propyl hydrogen (S)-pentylphosphonate, Esterase B | | Authors: | Colbert, D.A, Bennett, M.D, Lun, D.J, Holland, R, Delabre, M.-L, Loo, T.S, Anderson, B.F, Norris, G.E. | | Deposit date: | 2014-02-17 | | Release date: | 2014-08-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a C6-C4 SN3 inhibited ESTERASE B from LACTOBACILLUS RHAMNOSIS
TO BE PUBLISHED
|
|
5AAG
 
 | | Aurora A kinase bound to an imidazopyridine inhibitor (14b) | | Descriptor: | AURORA KINASE A, [3-[[4-[6-chloranyl-2-(1,3-dimethylpyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl]pyrazol-1-yl]methyl]phenyl]-(4-methylpiperazin-1-yl)methanone | | Authors: | McIntyre, P.J, Bayliss, R. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 7-(Pyrazol-4-Yl)-3H-Imidazo[4,5-B]Pyridine-Based Derivatives for Kinase Inhibition: Co-Crystallisation Studies with Aurora-A Reveal Distinct Differences in the Orientation of the Pyrazole N1-Substituent.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2X6E
 
 | | Aurora-A bound to an inhibitor | | Descriptor: | 6-BROMO-7-{4-[(5-METHYLISOXAZOL-3-YL)METHYL]PIPERAZIN-1-YL}-2-[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]-1H-IMIDAZO[4,5-B]PYRIDINE, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Imidazo[4,5-b]pyridine derivatives as inhibitors of Aurora kinases: lead optimization studies toward the identification of an orally bioavailable preclinical development candidate.
J. Med. Chem., 53, 2010
|
|
2X6D
 
 | | Aurora-A bound to an inhibitor | | Descriptor: | 6-BROMO-7-[4-(4-CHLOROBENZYL)PIPERAZIN-1-YL]-2-[4-(MORPHOLIN-4-YLMETHYL)PHENYL]-3H-IMIDAZO[4,5-B]PYRIDINE, SERINE/THREONINE-PROTEIN KINASE 6, SULFATE ION | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Imidazo[4,5-b]pyridine derivatives as inhibitors of Aurora kinases: lead optimization studies toward the identification of an orally bioavailable preclinical development candidate.
J. Med. Chem., 53, 2010
|
|
5GPB
 
 | |
6CQ1
 
 | | BCL6 BTB domain in complex with 15a | | Descriptor: | 2-{6-({[2-(1H-indol-3-yl)ethyl]carbamothioyl}amino)-3-[(4-methylpiperazin-1-yl)methyl]-1H-indol-1-yl}-N-(propan-2-yl)acetamide, B-cell lymphoma 6 protein | | Authors: | Linhares, B.M, Cheng, H, Xue, F, Cierpicki, T. | | Deposit date: | 2018-03-14 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69921041 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
6V4O
 
 | |
