7KKO
 
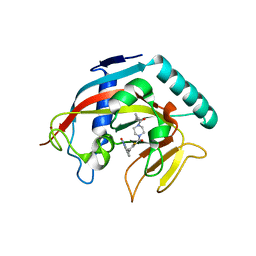 | | Structure of the catalytic domain of tankyrase 1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase, ZINC ION | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
3S6Y
 
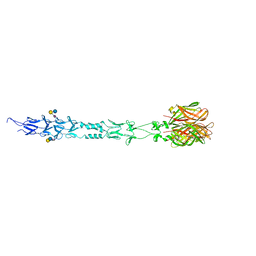 | | Structure of reovirus attachment protein sigma1 in complex with alpha-2,6-sialyllactose | | Descriptor: | N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Outer capsid protein sigma-1 | | Authors: | Reiter, D.M, Dermody, T.S, Stehle, T. | | Deposit date: | 2011-05-26 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of reovirus attachment protein sigma1 in complex with sialylated oligosaccharides
Plos Pathog., 7, 2011
|
|
7KKM
 
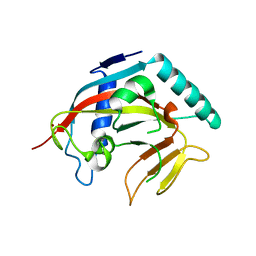 | | Structure of the catalytic domain of tankyrase 1 | | Descriptor: | Poly [ADP-ribose] polymerase, ZINC ION | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
8F24
 
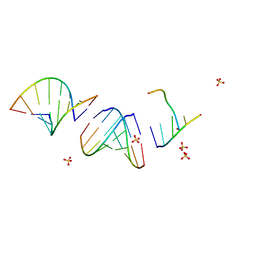 | | Mirror-image RNA octamer containing 2'-OMe-L-uridine | | Descriptor: | Mirror-image RNA 0G-XEC-0G-0U-0A-0C-0A-0C, SULFATE ION | | Authors: | Dantsu, Y, Zhang, W. | | Deposit date: | 2022-11-07 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Derivatization of Mirror-Image l-Nucleic Acids with 2'-OMe Modification for Thermal and Structural Stabilization.
Chembiochem, 24, 2023
|
|
7KKN
 
 | | Structure of the catalytic domain of tankyrase 1 in complex with talazoparib | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Poly [ADP-ribose] polymerase, ... | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
7KRF
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-4-(3-(2-cyanovinyl)-5-fluorophenoxy)-3-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenyl sulfurofluoridate (JLJ710) | | Descriptor: | 4-{3-[(E)-2-cyanoethenyl]-5-fluorophenoxy}-3-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenyl sulfurofluoridate, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Bertoletti, N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Covalent Inhibition of Wild-Type HIV-1 Reverse Transcriptase Using a Fluorosulfate Warhead.
Acs Med.Chem.Lett., 12, 2021
|
|
7KRC
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-4-(3-chloro-5-(2-cyanovinyl)phenoxy)-3-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenyl sulfurofluoridate (JLJ709) | | Descriptor: | 4-{3-chloro-5-[(E)-2-cyanoethenyl]phenoxy}-3-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenyl sulfurofluoridate, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Bertoletti, N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Covalent Inhibition of Wild-Type HIV-1 Reverse Transcriptase Using a Fluorosulfate Warhead.
Acs Med.Chem.Lett., 12, 2021
|
|
8W12
 
 | | Cryo-EM structure of VP3-VP6 heterohexamer | | Descriptor: | Core protein VP3, VP6 | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
8W1C
 
 | | Cryo-EM structure of BTV pre-subcore | | Descriptor: | Core protein VP3, VP6 | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
4O44
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in complex with 4-((4-(mesitylamino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ529), a non-nucleoside inhibitor | | Descriptor: | 4-({4-[3-(morpholin-4-yl)propoxy]-6-[(2,4,6-trimethylphenyl)amino]-1,3,5-triazin-2-yl}amino)benzonitrile, HIV-1 reverse transcriptase, p51 subunit, ... | | Authors: | Mislak, A.C, Frey, K.M, Anderson, K.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | A mechanistic and structural investigation of modified derivatives of the diaryltriazine class of NNRTIs targeting HIV-1 reverse transcriptase.
Biochim.Biophys.Acta, 1840, 2014
|
|
6LZ7
 
 | |
4O6E
 
 | | Discovery of 5,6,7,8-tetrahydropyrido[3,4-d]pyrimidine Inhibitors of Erk2 | | Descriptor: | Mitogen-activated protein kinase 1, N-[(1S)-1-(3-chloro-4-fluorophenyl)-2-hydroxyethyl]-2-(tetrahydro-2H-pyran-4-ylamino)-5,8-dihydropyrido[3,4-d]pyrimidine-7(6H)-carboxamide | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2013-12-20 | | Release date: | 2014-05-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of 5,6,7,8-tetrahydropyrido[3,4-d]pyrimidine inhibitors of Erk2.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1THN
 
 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA: inhibitory complex with ADP, crystal form I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigan, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-01 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
2FAZ
 
 | | Ubiquitin-Like Domain of Human Nuclear Zinc Finger Protein NP95 | | Descriptor: | Ubiquitin-like containing PHD and RING finger domains protein 1 | | Authors: | Walker, J.R, Wybenga-Groot, L, Doherty, R.S, Finerty Jr, P.J, Newman, E, Mackenzie, F.M, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-08 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ubiquitin-Like Domain of Human Nuclear Zinc Finger Protein NP95
To be Published
|
|
3QF9
 
 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a furan-thiazolidinedione ligand | | Descriptor: | 6-{5-[(Z)-(2,4-dioxo-1,3-thiazolidin-5-ylidene)methyl]furan-2-yl}-N-{3-[(4-ethylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl}naphthalene-1-carboxamide, CHLORIDE ION, Proto-oncogene serine/threonine-protein kinase pim-1, ... | | Authors: | Filippakopoulos, P, Bullock, A.N, Fedorov, O, Miduturu, C.V, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Grey, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-21 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a furan-thiazolidinedione ligand
To be Published
|
|
4A7C
 
 | | Crystal structure of PIM1 kinase with ETP46546 | | Descriptor: | ACETATE ION, IMIDAZOLE, N-(piperidin-4-ylmethyl)-3-[3-(trifluoromethyloxy)phenyl]-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | Authors: | Mazzorana, M, Montoya, G. | | Deposit date: | 2011-11-12 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Hit to Lead Evaluation of 1,2,3-Triazolo[4,5-B]Pyridines as Pim Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2QNQ
 
 | | HIV-1 Protease in complex with a chloro decorated pyrrolidine-based inhibitor | | Descriptor: | CHLORIDE ION, Gag-Pol polyprotein (Pr160Gag-Pol), N,N'-(3S,4S)-pyrrolidine-3,4-diylbis(N-benzyl-2-chlorobenzenesulfonamide) | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-07-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
6G99
 
 | | Solution structure of FUS-ZnF bound to UGGUG | | Descriptor: | RNA (5'-R(*UP*GP*GP*UP*G)-3'), RNA-binding protein FUS, ZINC ION | | Authors: | Loughlin, F.E, Allain, F.H.-T. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity.
Mol. Cell, 73, 2019
|
|
3WKV
 
 | | Voltage-gated proton channel: VSOP/Hv1 chimeric channel | | Descriptor: | Ion channel | | Authors: | Takeshita, K, Sakata, S, Yamashita, E, Fujiwara, Y, Kawanabe, A, Kurokawa, T, Okochi, Y, Matsuda, M, Narita, H, Okamura, Y, Nakagawa, A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.453 Å) | | Cite: | X-ray crystal structure of voltage-gated proton channel.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2D48
 
 | | Crystal structure of the Interleukin-4 variant T13D | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-11 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
1TIL
 
 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA:Poised for phosphorylation complex with ATP, crystal form II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigan, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
2O64
 
 | | Crystal structure of Pim1 with Quercetagetin | | Descriptor: | 3,5,6,7-TETRAHYDROXY-2-(3,4-DIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Holder, S, Zemskova, M, Zhang, C, Tabrizizad, M, Bremer, R, Neidigh, J.W, Lilly, M.B. | | Deposit date: | 2006-12-06 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase.
Mol.Cancer Ther., 6, 2007
|
|
3R01
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 5-bromo-7-methoxy-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Liu, J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2O3P
 
 | | Crystal structure of Pim1 with Quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Holder, S, Zemskova, M, Zhang, C, Tabrizizad, M, Bremer, R, Neidigh, J.W, Lilly, M.B. | | Deposit date: | 2006-12-01 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase.
Mol.Cancer Ther., 6, 2007
|
|
2O65
 
 | | Crystal structure of Pim1 with Pentahydroxyflavone | | Descriptor: | 5,7-DIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Holder, S, Zemskova, M, Zhang, C, Tabrizizad, M, Bremer, R, Neidigh, J.W, Lilly, M.B. | | Deposit date: | 2006-12-06 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase.
Mol.Cancer Ther., 6, 2007
|
|
