2YD8
 
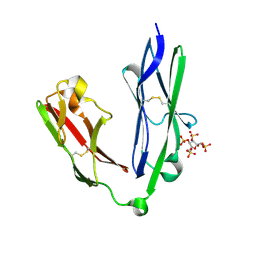 | | Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase LAR in complex with sucrose octasulphate | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE F | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-18 | | Release date: | 2011-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
2YU7
 
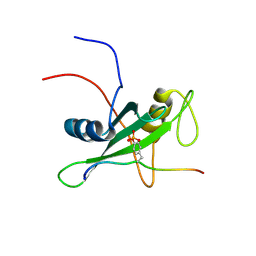 | | Solution structure of the SHP-1 C-terminal SH2 domain complexed with a tyrosine-phosphorylated peptide from NKG2A | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 6, natural killer group 2A | | Authors: | Kasai, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-15 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SHP-1 C-terminal SH2 domain complexed with a tyrosine-phosphorylated peptide from NKG2A
To be Published
|
|
6QFS
 
 | | Chargeless variant of the Cellulose-binding domain from Cellulomonas fimi | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ... | | Authors: | Young, D.R, Hoejgaard, C, Messens, J, Winther, J.R. | | Deposit date: | 2019-01-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Charge Interactions in a Highly Charge-depleted Protein
J.Am.Chem.Soc., 2021
|
|
4D01
 
 | | Crystal Structure of the Extracellular Domain of the Human Alpha9 Nicotinic Acetylcholine Receptor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-9 | | Authors: | Giastas, P, Zouridakis, M, Zarkadas, E, Tzartos, S.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Crystal Structures of Free and Antagonist-Bound States of Human Alpha9 Nicotinic Receptor Extracellular Domain
Nat.Struct.Mol.Biol., 21, 2014
|
|
5GI9
 
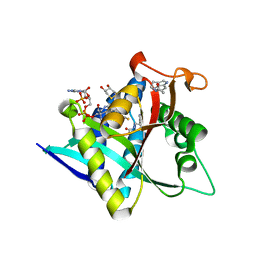 | | Crystal Structure of Drosophila melanogaster Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-Tryptamine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, COENZYME A, Dopamine N-acetyltransferase, ... | | Authors: | Yang, Y.C, Lin, S.J, Cheng, K.C, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2016-06-22 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An essential role of acetyl coenzyme A in the catalytic cycle of insect arylalkylamine N-acetyltransferase.
Commun Biol, 3, 2020
|
|
3NI2
 
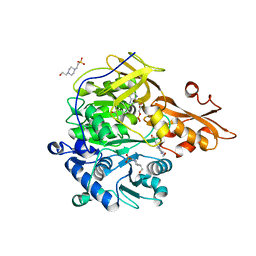 | | Crystal structures and enzymatic mechanisms of a Populus tomentosa 4-coumarate:CoA ligase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-coumarate:CoA ligase, 5'-O-{(S)-hydroxy[3-(4-hydroxyphenyl)propoxy]phosphoryl}adenosine | | Authors: | Hu, Y, Yin, L, Gai, Y, Wang, X.X, Wang, D.C. | | Deposit date: | 2010-06-14 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a Populus tomentosa 4-coumarate:CoA ligase shed light on its enzymatic mechanisms
Plant Cell, 22, 2010
|
|
5U20
 
 | | X-ray structure of the WlaRG aminotransferase from Campylobacter jejuni, internal PLP-aldimine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative aminotransferase, ... | | Authors: | Thoden, J.B, Holden, H.M, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
5U4N
 
 | |
5G5N
 
 | | Structure of the snake adenovirus 1 hexon-interlacing LH3 protein, methylmercury chloride derivative | | Descriptor: | CHLORIDE ION, GLYCEROL, LH3 HEXON-INTERLACING CAPSID PROTEIN, ... | | Authors: | Nguyen, T.H, Singh, A.K, Albala-Perez, B, van Raaij, M.J. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Reptilian Adenovirus Reveals a Phage Tailspike Fold Stabilizing a Vertebrate Virus Capsid.
Structure, 25, 2017
|
|
6EVF
 
 | | Structure of E285D S. cerevisiae Fdc1 with prFMN in the hydroxylated form | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, David, L, Payne, K.A.P. | | Deposit date: | 2017-11-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The role of conserved residues in Fdc decarboxylase in prenylated flavin mononucleotide oxidative maturation, cofactor isomerization, and catalysis.
J. Biol. Chem., 293, 2018
|
|
6QL9
 
 | | Structure of Fatty acid synthase complex from Saccharomyces cerevisiae at 2.9 Angstrom | | Descriptor: | 1,2-ETHANEDIOL, 4'-PHOSPHOPANTETHEINE, ACETATE ION, ... | | Authors: | Singh, K, Graf, B, Linden, A, Sautner, V, Urlaub, H, Tittmann, K, Stark, H, Chari, A. | | Deposit date: | 2019-01-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of a Regulatory Subunit of the Yeast Fatty Acid Synthase.
Cell, 180, 2020
|
|
3SXS
 
 | |
2R4U
 
 | | Crystal Structure of Wild-type E.coli GS in complex with ADP and Glucose(wtGSd) | | Descriptor: | (2R)-2-hydroxy-3-[4-(2-hydroxyethyl)piperazin-1-yl]propane-1-sulfonic acid, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sheng, F, Geiger, J. | | Deposit date: | 2007-09-01 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.367 Å) | | Cite: | The crystal structures of the open and catalytically competent closed conformation of Escherichia coli glycogen synthase.
J.Biol.Chem., 284, 2009
|
|
4HBV
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | Descriptor: | 1,2-ETHANEDIOL, 6-bromo-3-methyl-3,4-dihydroquinazolin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
9CRE
 
 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Alpha (B.1.1.7) variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Croll, T.I, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
2QIM
 
 | | Crystal Structure of Pathogenesis-related Protein LlPR-10.2B from yellow lupine in complex with Cytokinin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, CALCIUM ION, GLYCEROL, ... | | Authors: | Fernandes, H.C, Pasternak, O, Bujacz, G, Bujacz, A, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2007-07-05 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Lupinus luteus pathogenesis-related protein as a reservoir for cytokinin.
J.Mol.Biol., 378, 2008
|
|
9CRD
 
 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: B.1 variant 1 open RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Croll, T.I, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
6F5V
 
 | | Crystal structure of the prephenate aminotransferase from Arabidopsis thaliana | | Descriptor: | Bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase, CITRIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cobessi, D, Robin, A, Giustini, C, Graindorge, M, Matringe, M. | | Deposit date: | 2017-12-03 | | Release date: | 2019-03-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Febs J., 286, 2019
|
|
5U7R
 
 | | Identification of A New Class of Potent Cdc7 Inhibitors Designed by Putative Pharmacophore Model: Synthesis and Biological Evaluation of 2,3-Dihydrothieno[3,2-d]pyrimidin-4(1H)-ones | | Descriptor: | (1s,4s)-4-(4-fluorophenyl)-4-hydroxy-6'-(5-methyl-1H-pyrazol-4-yl)-1'H-spiro[cyclohexane-1,2'-thieno[3,2-d]pyrimidin]-4'(3'H)-one, Rho-associated protein kinase 2 | | Authors: | Hoffman, I.D, Skene, R.J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Identification of a new class of potent Cdc7 inhibitors designed by putative pharmacophore model: Synthesis and biological evaluation of 2,3-dihydrothieno[3,2-d]pyrimidin-4(1H)-ones.
Bioorg. Med. Chem., 25, 2017
|
|
7FRT
 
 | | PanDDA analysis group deposition of ground-state model of PTP1B, using pre-clustering, cluster 1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
1RP1
 
 | | DOG PANCREATIC LIPASE RELATED PROTEIN 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PANCREATIC LIPASE RELATED PROTEIN 1 | | Authors: | Roussel, A, Cambillau, C. | | Deposit date: | 1998-04-02 | | Release date: | 1998-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reactivation of the totally inactive pancreatic lipase RP1 by structure-predicted point mutations.
Proteins, 32, 1998
|
|
5DOM
 
 | | Crystal structure, maturation and flocculating properties of a 2S albumin from Moringa oleifera seeds | | Descriptor: | 1,2-ETHANEDIOL, 2S albumin, ACETATE ION | | Authors: | Ullah, A, Murakami, M.T, Arni, R.K. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of mature 2S albumin from Moringa oleifera seeds.
Biochem.Biophys.Res.Commun., 468, 2015
|
|
6L8S
 
 | | High resolution crystal structure of crustacean hemocyanin. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Masuda, T, Mikami, B, Baba, S. | | Deposit date: | 2019-11-07 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The high-resolution crystal structure of lobster hemocyanin shows its enzymatic capability as a phenoloxidase.
Arch.Biochem.Biophys., 688, 2020
|
|
6QR5
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 3-[1-[[4-(pyrrolidin-1-ylmethyl)phenyl]methyl]indol-6-yl]-1~{H}-pyrazol-5-amine, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6UMD
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-(pyridin-3-yl)-N-(5-{4-[(5-{[(pyridin-3-yl)acetyl]amino}-1,3,4-thiadiazol-2-yl)amino]piperidin-1-yl}-1,3,4-thiadiazol-2-yl)acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
