2W5I
 
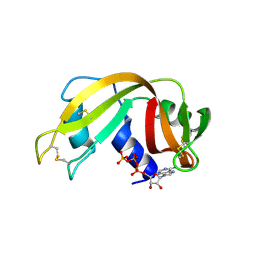 | | RNASE A-AP3A COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RIBONUCLEASE PANCREATIC | | Authors: | Chavali, G.B, Holloway, D.E, Baker, M.D, Acharya, K.R. | | Deposit date: | 2008-12-10 | | Release date: | 2009-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Influence of Naturally-Occurring 5'-Pyrophosphate-Linked Substituents on the Binding of Adenylic Inhibitors to Ribonuclease A: An X-Ray Crystallographic Study.
Biopolymers, 91, 2009
|
|
2PR4
 
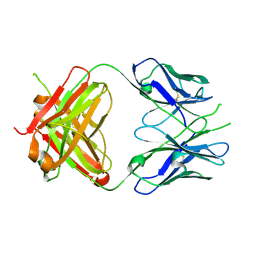 | | Crystal Structure of Fab' from the HIV-1 Neutralizing Antibody 2F5 | | Descriptor: | nmAb 2F5 Fab' Heavy Chain, nmAb 2F5 Fab' light Chain | | Authors: | Bryson, S, Julien, J.-P, Pai, E.F. | | Deposit date: | 2007-05-03 | | Release date: | 2007-05-15 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
3DLN
 
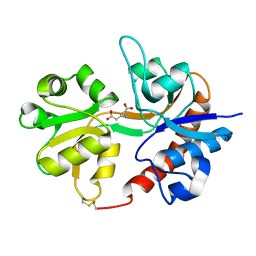 | | Crystal structure of the binding domain of the AMPA subunit GluR3 bound to glutamate | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Wang, Q, Sondermann, H, Oswald, R.E. | | Deposit date: | 2008-06-27 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of the S1S2 glutamate binding domain of GLuR3.
Proteins, 75, 2008
|
|
6D5O
 
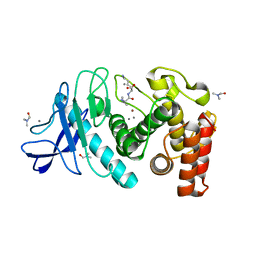 | | Hexagonal thermolysin (295 K) in the presence of 50% DMF | | Descriptor: | CALCIUM ION, DIMETHYLFORMAMIDE, LYSINE, ... | | Authors: | Juers, D.H. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.00005221 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5H8F
 
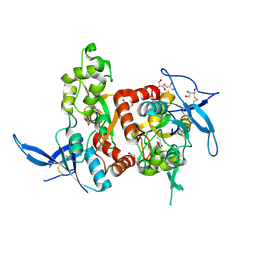 | | Structure of the apo human GluN1/GluN2A LBD | | Descriptor: | GLUTAMIC ACID, GLYCEROL, GLYCINE, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function.
Neuron, 89, 2016
|
|
5H8S
 
 | | Structure of the human GluA2 LBD in complex with GNE3419 | | Descriptor: | 7-[[ethyl(phenyl)amino]methyl]-2-methyl-[1,3,4]thiadiazolo[3,2-a]pyrimidin-5-one, CACODYLATE ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function.
Neuron, 89, 2016
|
|
9CT2
 
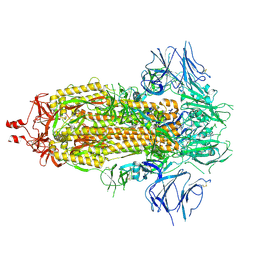 | | Cryo-EM structure of SARS-CoV-2 spike protein Ecto-domain with internal tag, All RBD down conformation, State-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Singh, S, Hasan, S.S. | | Deposit date: | 2024-07-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Production and cryo-electron microscopy structure of an internally tagged SARS-CoV-2 spike ecto-domain construct.
J Struct Biol X, 11, 2025
|
|
9CXE
 
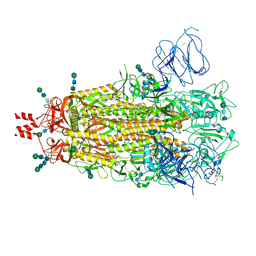 | | SARS CoV-2 Spike protein Ectodomain with internal tag, all RBD-down conformation -C1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, S, Hasan, S.S. | | Deposit date: | 2024-07-31 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Production and cryo-electron microscopy structure of an internally tagged SARS-CoV-2 spike ecto-domain construct.
J Struct Biol X, 11, 2025
|
|
5YCJ
 
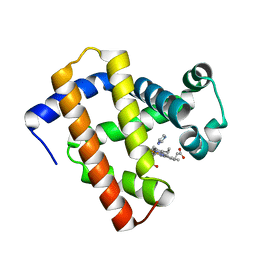 | | Ancestral myoglobin aMbWb' of Basilosaurus relative (polyphyly) imidazole-ligand | | Descriptor: | Ancestral myoglobin aMbWb' of Basilosaurus relative (polyphyly), IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Nakagawa, T, Tsuneshige, A, Shirai, T. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Tracing whale myoglobin evolution by resurrecting ancient proteins.
Sci Rep, 8, 2018
|
|
6D5P
 
 | |
9CVH
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein Ecto-domain with internal tag, 1RBD UP, State-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, S, Hasan, S.S. | | Deposit date: | 2024-07-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Production and cryo-electron microscopy structure of an internally tagged SARS-CoV-2 spike ecto-domain construct.
J Struct Biol X, 11, 2025
|
|
9CSS
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein Ecto-domain with internal tag, 1UP RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, S, Hasan, S.S. | | Deposit date: | 2024-07-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Production and cryo-electron microscopy structure of an internally tagged SARS-CoV-2 spike ecto-domain construct.
J Struct Biol X, 11, 2025
|
|
7YVC
 
 | | Aplysia californica FaNaC in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, Q.F, Liu, F.L, Dang, Y, Feng, H, Zhang, Z, Ye, S. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and mechanism of a neuropeptide-activated channel in the ENaC/DEG superfamily.
Nat.Chem.Biol., 19, 2023
|
|
1HL7
 
 | | Gamma lactamase from an Aureobacterium species in complex with 3a,4,7,7a-tetrahydro-benzo [1,3] dioxol-2-one | | Descriptor: | 3A,4,7,7A-TETRAHYDRO-BENZO [1,3] DIOXOL-2-ONE, GAMMA LACTAMASE | | Authors: | Line, K, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2003-03-14 | | Release date: | 2004-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structure of a (-)Gamma-Lactamase from an Aureobacterium Species Reveals a Tetrahedral Intermediate in the Active Site
J.Mol.Biol., 338, 2004
|
|
6DR4
 
 | |
5H8Q
 
 | | Structure of the human GluN1/GluN2A LBD in complex with GNE8324 | | Descriptor: | 6-[[ethyl-(4-fluorophenyl)amino]methyl]-2,3-dihydro-1~{H}-cyclopenta[3,4][1,3]thiazolo[1,4-~{a}]pyrimidin-8-one, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function.
Neuron, 89, 2016
|
|
3T4S
 
 | |
3LD6
 
 | | Crystal structure of human lanosterol 14alpha-demethylase (CYP51) in complex with ketoconazole | | Descriptor: | 1-acetyl-4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-imidazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazine, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Lanosterol 14-alpha demethylase, ... | | Authors: | Strushkevich, N, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-12 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of human CYP51 inhibition by antifungal azoles.
J.Mol.Biol., 397, 2010
|
|
1L7G
 
 | | Crystal structure of E119G mutant influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
3AKA
 
 | | Structural basis for prokaryotic calcium-mediated regulation by a Streptomyces coelicolor calcium-binding protein | | Descriptor: | CALCIUM ION, Putative calcium binding protein | | Authors: | Zhao, X, Pang, H, Wang, S, Zhou, W, Yang, K, Bartlam, M. | | Deposit date: | 2010-07-09 | | Release date: | 2011-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for prokaryotic calciummediated regulation by a Streptomyces coelicolor calcium binding protein
Protein Cell, 1, 2010
|
|
6D5T
 
 | | Hexagonal thermolysin cryocooled to 100 K with 50% MPD as cryoprotectant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, LYSINE, ... | | Authors: | Juers, D.H. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.00003719 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6Y6D
 
 | | Tubulin-7-Aminonoscapine complex | | Descriptor: | (3~{S})-7-azanyl-6-methoxy-3-[(5~{R})-4-methoxy-6-methyl-7,8-dihydro-5~{H}-[1,3]dioxolo[4,5-g]isoquinolin-5-yl]-3~{H}-2-benzofuran-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Oliva, M.A, Prota, A.E, Rodriguez-Salarichs, J, Gu, W, Bennani, Y.L, Jimenez-Barbero, J, Canales, A, Steinmetz, M.O, Diaz, J.F. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Noscapine Activation for Tubulin Binding.
J.Med.Chem., 63, 2020
|
|
7B6J
 
 | | Crystal structure of MurE from E.coli in complex with minifrag succinimide | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
4JVR
 
 | | Co-crystal structure of MDM2 with inhibitor (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide | | Descriptor: | (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
7B68
 
 | | Crystal structure of MurE from E.coli in complex with Z57299526 | | Descriptor: | 4-[(4-methylphenyl)methyl]-1,4-thiazinane 1,1-dioxide, DIMETHYL SULFOXIDE, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
