7NPF
 
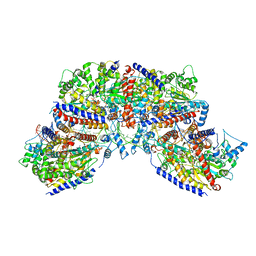 | | Vibrio cholerae ParA2-ATPyS-DNA filament | | Descriptor: | AAA family ATPase, DNA (49-MER), MAGNESIUM ION, ... | | Authors: | Parker, A.V, Bergeron, J.R.C. | | Deposit date: | 2021-02-26 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The structure of the bacterial DNA segregation ATPase filament reveals the conformational plasticity of ParA upon DNA binding.
Nat Commun, 12, 2021
|
|
5IHJ
 
 | |
2IJ0
 
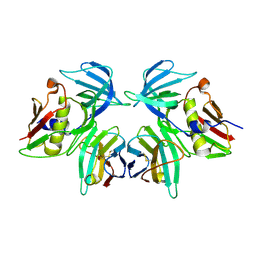 | | Structural basis of T cell specificity and activation by the bacterial superantigen toxic shock syndrome toxin-1 | | Descriptor: | Toxic shock syndrome toxin-1, penultimate affinity-matured variant of hVbeta 2.1, D10 | | Authors: | Moza, B, Varma, A.K, Buonpane, R.A, Zhu, P, Herfst, C.A, Nicholson, M.J, Wilbuer, A.K, Nulifer, S, Wucherpfenning, K.W, McCormick, J.K, Kranz, D.M, Sundberg, E.J. | | Deposit date: | 2006-09-28 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of T-cell specificity and activation by the bacterial superantigen TSST-1.
Embo J., 26, 2007
|
|
4C5Z
 
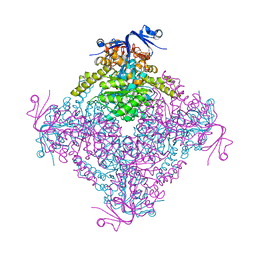 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
8PMJ
 
 | | Vanadate-trapped BSEP in nanodiscs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Bile salt export pump, ... | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
8PMD
 
 | | Nucleotide-bound BSEP in nanodiscs | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bile salt export pump, MAGNESIUM ION | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
5J7A
 
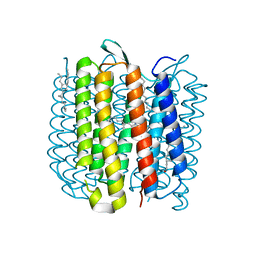 | | Bacteriorhodopsin ground state structure obtained with Serial Femtosecond Crystallography | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Nogly, P, Panneels, V, Nelson, G, Gati, C, Kimura, T, Milne, C, Milathianaki, D, Kubo, M, Wu, W, Conrad, C, Coe, J, Bean, R, Zhao, Y, Bath, P, Dods, R, Harimoorthy, R, Beyerlein, K.R, Rheinberger, J, James, D, DePonte, D, Li, C, Sala, L, Williams, G, Hunter, M, Koglin, J.E, Berntsen, P, Nango, E, Iwata, S, Chapman, H.N, Fromme, P, Frank, M, Abela, R, Boutet, S, Barty, A, White, T.A, Weierstall, U, Spence, J, Neutze, R, Schertler, G, Standfuss, J. | | Deposit date: | 2016-04-06 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lipidic cubic phase injector is a viable crystal delivery system for time-resolved serial crystallography.
Nat Commun, 7, 2016
|
|
3J1X
 
 | | A refined model of the prototypical Salmonella typhimurium T3SS basal body reveals the molecular basis for its assembly | | Descriptor: | Protein PrgH | | Authors: | Sgourakis, N.G, Bergeron, J.R.C, Worrall, L.J, Strynadka, N.C.J, Baker, D. | | Deposit date: | 2012-07-10 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | A Refined Model of the Prototypical Salmonella SPI-1 T3SS Basal Body Reveals the Molecular Basis for Its Assembly.
Plos Pathog., 9, 2013
|
|
2IY3
 
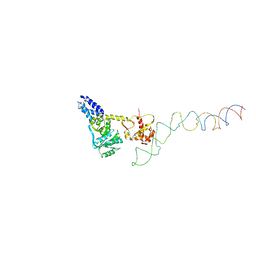 | | Structure of the E. Coli Signal Regognition Particle | | Descriptor: | 4.5S RNA, SIGNAL SEQUENCE, Signal recognition particle protein,Signal recognition particle 54 kDa protein | | Authors: | Schaffitzel, C, Oswald, M, Berger, I, Ishikawa, T, Abrahams, J.P, Koerten, H.K, Koning, R.I, Ban, N. | | Deposit date: | 2006-07-12 | | Release date: | 2006-11-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structure of the E. Coli Signal Recognition Particle Bound to a Translating Ribosome
Nature, 444, 2006
|
|
8QRO
 
 | | ASCT2 trimer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRP
 
 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.1) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRQ
 
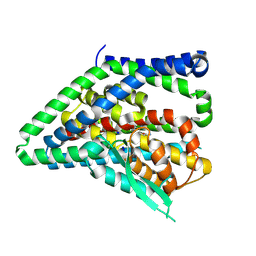 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.2) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRR
 
 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.3) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRS
 
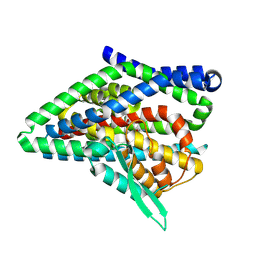 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the intermediate outward-facing state (iOFS-up) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRU
 
 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the intermediate outward-facing state (iOFS-down) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRV
 
 | | ASCT2 protomer in lipid nanodiscs under low Na+ concentration in the outward-facing state (OFS) | | Descriptor: | Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRW
 
 | | ASCT2 protomer in lipid nanodiscs under low Na+ concentration in the intermediate outward-facing state (iOFS-up) | | Descriptor: | Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
2O5D
 
 | |
8AS5
 
 | |
8PM6
 
 | | Human bile salt export pump (BSEP) in complex with inhibitor GBM in nanodiscs | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Bile salt export pump, CHOLESTEROL | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
5KEI
 
 | |
3J1V
 
 | |
3JCS
 
 | | 2.8 Angstrom cryo-EM structure of the large ribosomal subunit from the eukaryotic parasite Leishmania | | Descriptor: | 26S alpha ribosomal RNA, 26S delta ribosomal RNA, 26S epsilon ribosomal RNA, ... | | Authors: | Shalev-Benami, M, Zhang, Y, Matzov, D, Halfon, Y, Zackay, A, Rozenberg, H, Zimmerman, E, Bashan, A, Jaffe, C.L, Yonath, A, Skiniotis, G. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-20 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | 2.8- angstrom Cryo-EM Structure of the Large Ribosomal Subunit from the Eukaryotic Parasite Leishmania.
Cell Rep, 16, 2016
|
|
1S4X
 
 | | NMR Structure of the integrin B3 cytoplasmic domain in DPC micelles | | Descriptor: | Integrin beta-3 | | Authors: | Vinogradova, O, Vaynberg, J, Kong, X, Haas, T.A, Plow, E.F, Qin, J. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Membrane-mediated structural transitions at the cytoplasmic face during integrin activation.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3KOU
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD38 molecule, ... | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Oppenheimer, N.J, Kellenberger, E, Schuber, F. | | Deposit date: | 2009-11-13 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
