9CCX
 
 | | Crystal Structure of the Klebsiella pneumoniae LpxH/JH-LPH-86 complex | | Descriptor: | 1-(5-{4-[4-(trifluoromethyl)pyridin-2-yl]piperazine-1-sulfonyl}-2,3-dihydro-1H-indol-1-yl)ethan-1-one, MANGANESE (II) ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Cochrane, C.S, Zhou, P. | | Deposit date: | 2024-06-23 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Evaluation of Pyridinyl Sulfonyl Piperazine LpxH Inhibitors with Potent Antibiotic Activity Against Enterobacterales.
Jacs Au, 4, 2024
|
|
9CCY
 
 | | Crystal structure of the Klebsiella pneumoniae LpxH / JH-LPH-90 complex | | Descriptor: | 1-(5-{4-[6-(trifluoromethyl)pyridin-2-yl]piperazine-1-sulfonyl}-2,3-dihydro-1H-indol-1-yl)ethan-1-one, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Cochrane, C.S, Zhou, P. | | Deposit date: | 2024-06-23 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design and Evaluation of Pyridinyl Sulfonyl Piperazine LpxH Inhibitors with Potent Antibiotic Activity Against Enterobacterales.
Jacs Au, 4, 2024
|
|
4MTR
 
 | | Zn-bound GloA2 | | Descriptor: | 1,2-ETHANEDIOL, Lactoylglutathione lyase, ZINC ION | | Authors: | Bythell-Douglas, R, Bond, C.S. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of a homodimeric Pseudomonas glyoxalase I enzyme reveals asymmetric metallation commensurate with half-of-sites activity.
Chemistry, 21, 2015
|
|
8X54
 
 | | Cryo-EM structure of human gamma-secretase in complex with APP-C99 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-11-16 | | Release date: | 2024-05-29 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism of substrate recognition and cleavage by human gamma-secretase.
Science, 384, 2024
|
|
9GPC
 
 | |
8X52
 
 | | Cryo-EM structure of human gamma-secretase in complex with Abeta49 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-11-16 | | Release date: | 2024-05-29 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism of substrate recognition and cleavage by human gamma-secretase.
Science, 384, 2024
|
|
5DT2
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD11 [N4-methyl-N2-(2-methyl-1-(2-phenoxyphenyl)-1H-indol-6-yl)pyrimidine-2,4-diamine] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~4~-methyl-N~2~-[2-methyl-1-(2-phenoxyphenyl)-1H-indol-6-yl]pyrimidine-2,4-diamine, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-17 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
8OQZ
 
 | | Structure of human gamma-secretase PSEN1 APH-1B isoform reconstituted into lipid nanodisc in complex with Ab46 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Odorcic, I, Chavez Gutierrez, L, Efremov, R.G. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Apo and A beta 46-bound gamma-secretase structures provide insights into amyloid-beta processing by the APH-1B isoform.
Nat Commun, 15, 2024
|
|
6GB0
 
 | |
7KZA
 
 | |
4NA9
 
 | | Factor VIIa in complex with the inhibitor 3'-amino-5'-[(2s,4r)-6-carbamimidoyl-4-phenyl-1,2,3,4-tetrahydroquinolin-2-yl]biphenyl-2-carboxylic acid | | Descriptor: | 3'-amino-5'-[(2S,4R)-6-carbamimidoyl-4-phenyl-1,2,3,4-tetrahydroquinolin-2-yl]biphenyl-2-carboxylic acid, CALCIUM ION, Coagulation factor VII heavy chain, ... | | Authors: | Wei, A. | | Deposit date: | 2013-10-21 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Tetrahydroquinoline Derivatives as Potent and Selective Factor XIa Inhibitors.
J.Med.Chem., 57, 2014
|
|
8X53
 
 | | Cryo-EM structure of human gamma-secretase in complex with Abeta46 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-11-16 | | Release date: | 2024-05-29 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular mechanism of substrate recognition and cleavage by human gamma-secretase.
Science, 384, 2024
|
|
6BFG
 
 | |
5TDX
 
 | | Resurrected Ancestral Hydroxynitrile Lyase from Flowering Plants | | Descriptor: | Ancestral Hydroxynitrile Lyase 1, GLYCEROL | | Authors: | Jones, B.J, Evans, R, Wilmot, C.M, Kazlauskas, R.J. | | Deposit date: | 2016-09-20 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Larger active site in an ancestral hydroxynitrile lyase increases catalytically promiscuous esterase activity.
Plos One, 15, 2020
|
|
3ZUC
 
 | | Structure of CBM3b of major scaffoldin subunit ScaA from Acetivibrio cellulolyticus determined from the crystals grown in the presence of Nickel | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDIN, ... | | Authors: | Yaniv, O, Halfon, Y, Lamed, R, Frolow, F. | | Deposit date: | 2011-07-18 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.001 Å) | | Cite: | Structure of Cbm3B of the Major Scaffoldin Subunit Scaa from Acetivibrio Cellulolyticus
Acta Crystallogr.,Sect.F, 68, 2012
|
|
8C87
 
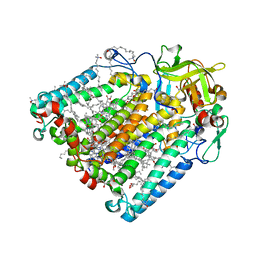 | | Double mutant A(L172)C/L(L246)C structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-ETHANEDIOL, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A, Selikhanov, G, Fufina, T, Vasilieva, L, Atamas, A, Yukhimchuk, D. | | Deposit date: | 2023-01-19 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds.
Membranes (Basel), 13, 2023
|
|
6FPY
 
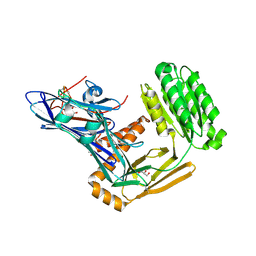 | | Inter-alpha-inhibitor heavy chain 1, wild type | | Descriptor: | GLYCEROL, Inter-alpha-trypsin inhibitor heavy chain H1, MAGNESIUM ION | | Authors: | Briggs, D.C, Day, A.J. | | Deposit date: | 2018-02-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Inter-alpha-inhibitor heavy chain-1 has an integrin-like 3D structure mediating immune regulatory activities and matrix stabilization during ovulation
J.Biol.Chem., 2020
|
|
2VBE
 
 | | Tailspike protein of bacteriophage Sf6 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Barbirz, S, Mueller, J.J, Freiberg, A, Heinemann, U, Seckler, R. | | Deposit date: | 2007-09-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An intersubunit active site between supercoiled parallel beta helices in the trimeric tailspike endorhamnosidase of Shigella flexneri Phage Sf6.
Structure, 16, 2008
|
|
6GJN
 
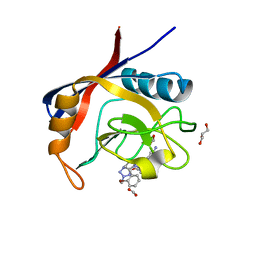 | | Cyclophilin A complexed with tri-vector ligand 15. | | Descriptor: | 1-[(4-aminophenyl)methyl]-3-[2-[(2~{R})-2-(2-bromophenyl)pyrrolidin-1-yl]-2-oxidanylidene-ethyl]-1-[(2-methyl-1,2,3,4-tetrazol-5-yl)methyl]urea, FORMIC ACID, GLYCEROL, ... | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-16 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
6G25
 
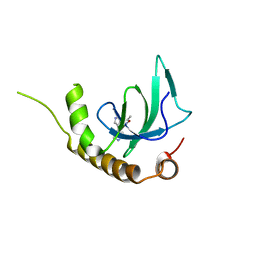 | | X-ray structure of NSD3-PWWP1 in complex with compound 4 | | Descriptor: | 3,5-dimethyl-4-(4-pyridin-4-yl-1~{H}-pyrazol-3-yl)-1,2-oxazole, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
7UY1
 
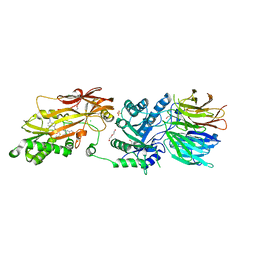 | | HUMAN PRMT5:MEP50 COMPLEX WITH MTA and Fragment 5 Bound | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-1,5-naphthyridin-2-amine, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Gunn, R.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
6GNT
 
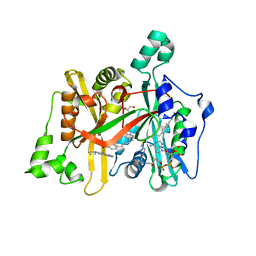 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and a Quionlinyl aryl sulphonamide ligand | | Descriptor: | 4-[4-(1-methylpiperidin-4-yl)butyl]-~{N}-(6-pyrrolidin-1-ylquinolin-5-yl)benzenesulfonamide, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, ... | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
8HBF
 
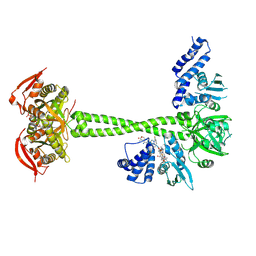 | |
8XNF
 
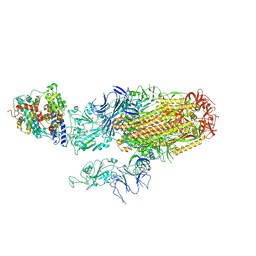 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P) in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
5G3P
 
 | | Bacillus cereus formamidase (BceAmiF) acetylated at the active site. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Conejero-Muriel, M, Martinez-Rodriguez, S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A novel cysteine carbamoyl-switch is responsible for the inhibition of formamidase, a nitrilase superfamily member.
Arch.Biochem.Biophys., 662, 2019
|
|
