6AAU
 
 | | Solution Structure for m62A helix 45 in 3' end of 12S rRNA | | Descriptor: | RNA (24-mer) | | Authors: | Liu, X, Wu, P. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
5II0
 
 | |
5I9T
 
 | | Caspase 3 V266C | | Descriptor: | ACE-ASP-GLU-VAL-ASK, ACETATE ION, Caspase-3 | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-20 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3EWY
 
 | | K314A mutant of human orotidyl-5'-monophosphate decarboxylase soaked with OMP, decarboxylated to UMP | | Descriptor: | GLYCEROL, Orotidine-5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Heinrich, D, Diederichsen, U, Rudolph, M. | | Deposit date: | 2008-10-16 | | Release date: | 2009-04-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase
Chemistry, 15, 2009
|
|
6AC8
 
 | | Crystal structure of Mycobacterium smegmatis Mfd at 2.75 A resolution | | Descriptor: | Mycobacterium smegmatis Mfd, SULFATE ION | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
8TVY
 
 | | Cryo-EM structure of CPD lesion containing RNA Polymerase II elongation complex with Rad26 and Elf1 (closed state) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3EX1
 
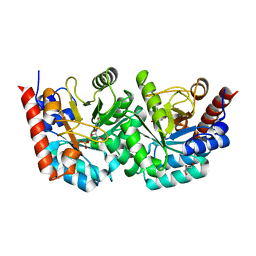 | | human orotidyl-5'-monophosphate decarboxylase soaked with 6-cyano-UMP, converted to UMP | | Descriptor: | 6-cyanouridine 5'-phosphate, GLYCEROL, Orotidine-5'-phosphate decarboxylase, ... | | Authors: | Heinrich, D, Diederichsen, U, Rudolph, M. | | Deposit date: | 2008-10-16 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase
Chemistry, 15, 2009
|
|
3EXD
 
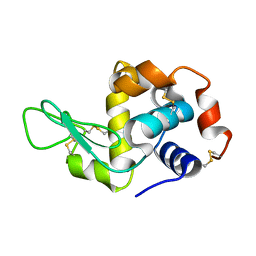 | |
3IX1
 
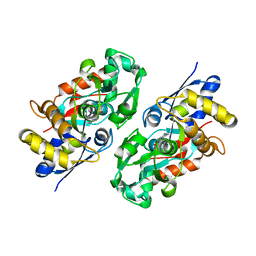 | | Periplasmic N-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding protein from Bacillus halodurans | | Descriptor: | N-[(4-amino-2-methylpyrimidin-5-yl)methyl]formamide, N-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding protein | | Authors: | Bale, S, Rajashankar, K.R, Perry, K, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-09-03 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | HMP Binding Protein ThiY and HMP-P Synthase THI5 Are Structural Homologues.
Biochemistry, 49, 2010
|
|
5LIG
 
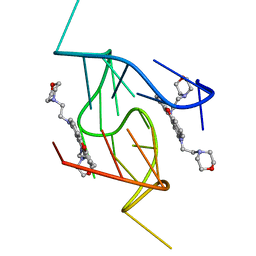 | | G-Quadruplex formed at the 5'-end of NHEIII_1 Element in human c-MYC promoter bound to triangulenium based fluorescence probe DAOTA-M2 | | Descriptor: | 8,12-bis(2-morpholinoethyl)-8H-benzo[ij]xantheno[1,9,8-cdef][2,7]naphthyridin-12-iumhexafluorophosphate, DNA (5'-D(*TP*AP*GP*GP*GP*AP*GP*GP*GP*TP*AP*GP*GP*GP*AP*GP*GP*GP*T)-3') | | Authors: | Kotar, A, Wang, B, Shivalingam, A, Gonzalez-Garcia, J, Vilar, R, Plavec, J. | | Deposit date: | 2016-07-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Triangulenium-Based Long-Lived Fluorescence Probe Bound to a G-Quadruplex.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3EX2
 
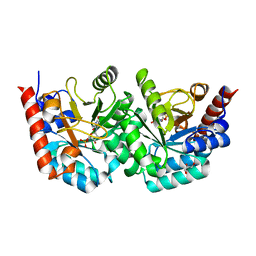 | |
3EWW
 
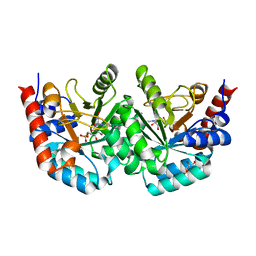 | | D312N mutant of human orotidyl-5'-monophosphate decarboxylase in complex with 6-cyano-UMP, covalent adduct | | Descriptor: | 6-[(E)-iminomethyl]uridine 5'-phosphate, GLYCEROL, Orotidine-5'-phosphate decarboxylase | | Authors: | Heinrich, D, Wittmann, J, Diederichsen, U, Rudolph, M. | | Deposit date: | 2008-10-16 | | Release date: | 2009-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase
Chemistry, 15, 2009
|
|
5IP7
 
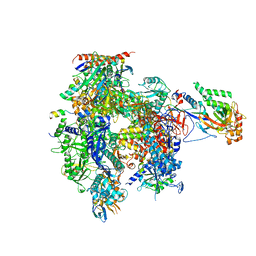 | | Structure of RNA Polymerase II-Tfg1 peptide complex | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Transcription initiation complex structures elucidate DNA opening.
Nature, 533, 2016
|
|
3EWZ
 
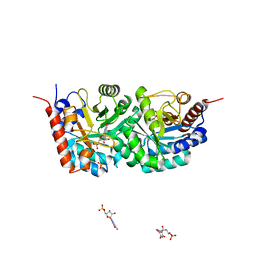 | |
2MOW
 
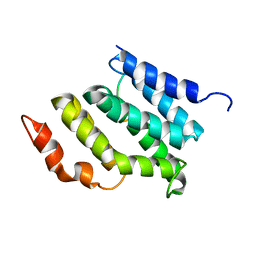 | |
5IPS
 
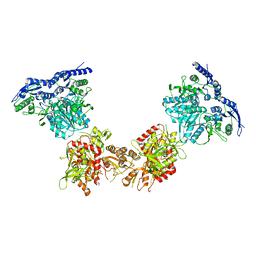 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 4 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5IRI
 
 | |
6A16
 
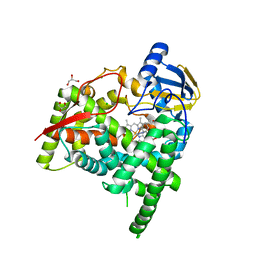 | | Crystal structure of CYP90B1 in complex with uniconazole | | Descriptor: | (1E,3S)-1-(4-chlorophenyl)-4,4-dimethyl-2-(1H-1,2,4-triazol-1-yl)pent-1-en-3-ol, CHLORIDE ION, Cytochrome P450 90B1, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
7M3T
 
 | |
1FMV
 
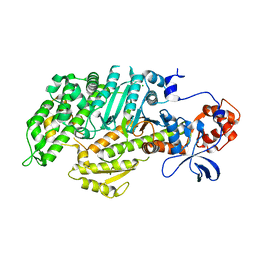 | | CRYSTAL STRUCTURE OF THE APO MOTOR DOMAIN OF DICTYOSTELLIUM MYOSIN II | | Descriptor: | CHLORIDE ION, MYOSIN II HEAVY CHAIN | | Authors: | Bauer, C.B, Holden, H.M, Thoden, J.B, Smith, R, Rayment, I. | | Deposit date: | 2000-08-18 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structures of the apo and MgATP-bound states of Dictyostelium discoideum myosin motor domain.
J.Biol.Chem., 275, 2000
|
|
5I9W
 
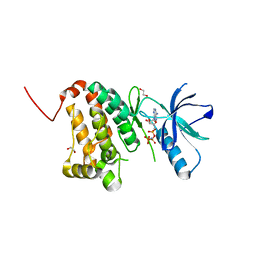 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with ANP | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.359 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
5I3Q
 
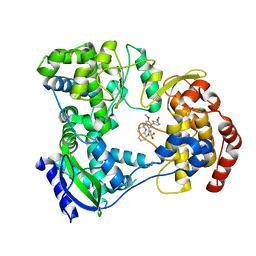 | | DENGUE SEROTYPE 3 RNA-DEPENDENT RNA POLYMERASE BOUND TO COMPOUND 29 | | Descriptor: | 5-[5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl]-4-methoxy-2-methyl-N-[(quinolin-8-yl)sulfonyl]benzamide, Genome polyprotein, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-02-10 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling
Plos Pathog., 12, 2016
|
|
5IA5
 
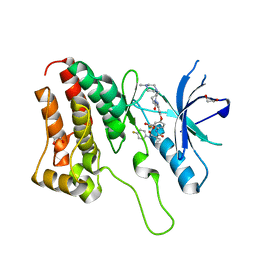 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with golvatinib (E7050) | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, golvatinib | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
5IAR
 
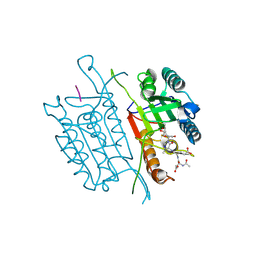 | | Caspase 3 V266W | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACE-ASP-GLU-VAL-ASK, Caspase-3, ... | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-21 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8AF7
 
 | | Room temperature SSX crystal structure of CTX-M-14 (10K dataset) | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Oberthuer, D, Perbandt, M, Prester, A, Rohde, H, Betzel, C, Yefanov, O. | | Deposit date: | 2022-07-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
