7CBH
 
 | | Crystal structure of threonyl-tRNA synthetase (ThrRS) from Salmonella enterica in complex with an inhibitor | | Descriptor: | Threonine--tRNA ligase, ZINC ION, [(E)-4-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)but-2-enyl] (2S,3R)-2-azanyl-3-oxidanyl-butanoate | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2020-06-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided optimization and mechanistic study of a class of quinazolinone-threonine hybrids as antibacterial ThrRS inhibitors.
Eur.J.Med.Chem., 207, 2020
|
|
7LNU
 
 | | Ternary complex of the Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus bound to isopentenyl monophosphate and ATP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
7LNW
 
 | |
7LNT
 
 | | Ternary complex of the Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus bound to benzyl monophosphate and ATP | | Descriptor: | (phenylmethyl) dihydrogen phosphate, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
7V9D
 
 | | Plasmodium falciparum Prolyl-tRNA Synthetase (PfPRS) in Complex with inhibitor L95 and azetidine | | Descriptor: | (2S)-azetidine-2-carboxylic acid, 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, ... | | Authors: | Manickam, Y, Malhotra, N, Sharma, A. | | Deposit date: | 2021-08-24 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Plasmodium falciparum Prolyl-tRNA Synthetase (PfPRS) in Complex with inhibitor L95 and azetidine
To Be Published
|
|
7LNV
 
 | | Apo Structure of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus | | Descriptor: | GLYCEROL, Isopentenyl phosphate kinase, MALONATE ION | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
8PIY
 
 | |
5DFD
 
 | | Crystal structure of BRD2(BD2) W370F mutant with ligand 28 bound | | Descriptor: | Bromodomain-containing protein 2, GLYCEROL, methyl [(4S)-6-(1H-indol-4-yl)-8-methoxy-1-methyl-4H-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]acetate | | Authors: | Tallant, C, Baud, M, Lin-Shiao, E, Chirgadze, D.Y, Ciulli, A. | | Deposit date: | 2015-08-26 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New Synthetic Routes to Triazolo-benzodiazepine Analogues: Expanding the Scope of the Bump-and-Hole Approach for Selective Bromo and Extra-Terminal (BET) Bromodomain Inhibition.
J.Med.Chem., 59, 2016
|
|
5E4N
 
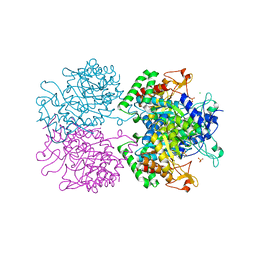 | | 3-Deoxy-D-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis with D-tyrosine bound in the tyrosine and phenylalanine binding sites | | Descriptor: | 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase, CHLORIDE ION, D-TYROSINE, ... | | Authors: | Reichau, S, Jiao, W, Parker, E.J. | | Deposit date: | 2015-10-06 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Probing the Sophisticated Synergistic Allosteric Regulation of Aromatic Amino Acid Biosynthesis in Mycobacterium tuberculosis Using -Amino Acids.
Plos One, 11, 2016
|
|
1MX7
 
 | |
3KQ4
 
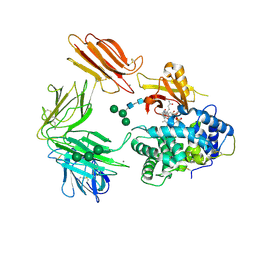 | | Structure of Intrinsic Factor-Cobalamin bound to its receptor Cubilin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Andersen, C.B.F, Madsen, M, Moestrup, S.K, Andersen, G.R. | | Deposit date: | 2009-11-17 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for receptor recognition of vitamin-B(12)-intrinsic factor complexes.
Nature, 464, 2010
|
|
6KZZ
 
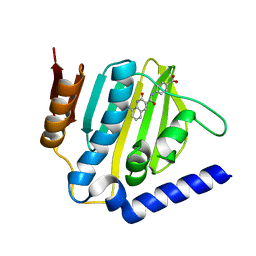 | | Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 4-[[8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]benzoic acid, DNA gyrase subunit B | | Authors: | Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.
Acs Omega, 5, 2020
|
|
7SA5
 
 | | Two-state solution NMR structure of Apo Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Born, A, Vogeli, B. | | Deposit date: | 2021-09-22 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Reconstruction of Coupled Intra- and Interdomain Protein Motion from Nuclear and Electron Magnetic Resonance.
J.Am.Chem.Soc., 143, 2021
|
|
7YXU
 
 | | Crystal structure of agonistic antibody 1618 fab domain bound to human 4-1BB. | | Descriptor: | MANGANESE (II) ION, Tumor necrosis factor receptor superfamily member 9, heavy chain of Fab, ... | | Authors: | Hakansson, M, Rose, N, Petersson, J, Enell Smith, K, Thorolfsson, M, von Schantz, L. | | Deposit date: | 2022-02-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Bispecific Tumor Antigen-Conditional 4-1BB x 5T4 Agonist, ALG.APV-527, Mediates Strong T-Cell Activation and Potent Antitumor Activity in Preclinical Studies.
Mol.Cancer Ther., 22, 2023
|
|
4ALX
 
 | | Crystal Structure of Ls-AChBP complexed with the potent nAChR antagonist DHbE | | Descriptor: | (4bS,6S)-6-methoxy-1,4,6,7,9,10,12,13-octahydro-3H,5H-pyrano[4',3':3,4]pyrido[2,1-i]indol-3-one, ACETYLCHOLINE BINDING PROTEIN, MAGNESIUM ION, ... | | Authors: | Shahsavar, A, Kastrup, J.S, Nielsen, E.O, Kristensen, J.L, Gajhede, M, Balle, T. | | Deposit date: | 2012-03-06 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Lymnaea Stagnalis Achbp Complexed with the Potent Nachr Antagonist Dh-Betab-E Suggests a Unique Mode of Antagonism
Plos One, 7, 2012
|
|
2IMG
 
 | | Crystal structure of dual specificity protein phosphatase 23 from Homo sapiens in complex with ligand malate ion | | Descriptor: | D-MALATE, Dual specificity protein phosphatase 23 | | Authors: | Agarwal, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of human dual specificity protein phosphatase 23, VHZ, enzyme-substrate/product complex.
J.Biol.Chem., 283, 2008
|
|
3DQB
 
 | | Crystal structure of the active G-protein-coupled receptor opsin in complex with a C-terminal peptide derived from the Galpha subunit of transducin | | Descriptor: | 11meric peptide form Guanine nucleotide-binding protein G(t) subunit alpha-1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Scheerer, P, Park, J.H, Hildebrand, P.W, Kim, Y.J, Krauss, N, Choe, H.-W, Hofmann, K.P, Ernst, O.P. | | Deposit date: | 2008-07-09 | | Release date: | 2008-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of opsin in its G-protein-interacting conformation
Nature, 455, 2008
|
|
4BAM
 
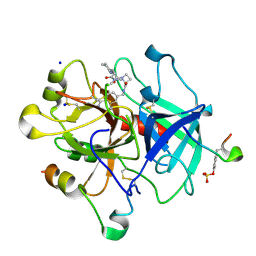 | | Thrombin in complex with inhibitor | | Descriptor: | (2S)-N-[(4-carbamimidoylphenyl)methyl]-1-[(2R)-2-cyclohexyl-2-[[2-(dimethylamino)-2-oxidanylidene-ethyl]amino]ethanoyl]azetidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN VARIANT-1, ... | | Authors: | Xue, Y, Musil, D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Identification of Structure-Kinetic and Structure-Thermodynamic Relationships for Thrombin Inhibitors.
Biochemistry, 52, 2013
|
|
2WOH
 
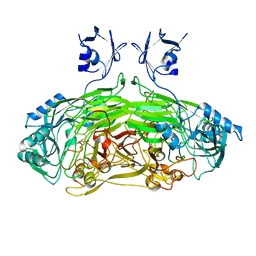 | | Strontium soaked E. coli copper amine oxidase | | Descriptor: | CALCIUM ION, COPPER (II) ION, PRIMARY AMINE OXIDASE, ... | | Authors: | Smith, M.A, Pirrat, P, Pearson, A.R, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2009-07-23 | | Release date: | 2010-05-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Exploring the Roles of the Metal Ions in Escherichia Coli Copper Amine Oxidase.
Biochemistry, 49, 2010
|
|
2QSU
 
 | |
4BAH
 
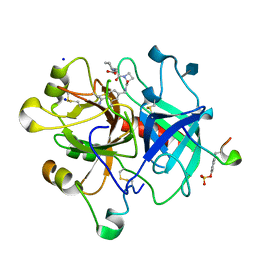 | | Thrombin in complex with inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN VARIANT-1, SODIUM ION, ... | | Authors: | Xue, Y, Musil, D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification of Structure-Kinetic and Structure-Thermodynamic Relationships for Thrombin Inhibitors.
Biochemistry, 52, 2013
|
|
8VLB
 
 | | Crystal structure of EloBC-VHL-CDO1 complex bound to compound 4 molecular glue | | Descriptor: | CITRIC ACID, Cysteine dioxygenase type 1, Elongin-B, ... | | Authors: | Shu, W, Ma, X, Tutter, A, Buckley, D, Golosov, A, Michaud, G. | | Deposit date: | 2024-01-11 | | Release date: | 2024-03-13 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A small-molecule VHL molecular glue degrader for cysteine dioxygenase 1.
Nat.Chem.Biol., 2025
|
|
4BAQ
 
 | | Thrombin in complex with inhibitor | | Descriptor: | (2S)-N-[(4-carbamimidoylphenyl)methyl]-1-[(2R)-2-cyclohexyl-2-[[2-(ethylamino)-2-oxidanylidene-ethyl]amino]ethanoyl]azetidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN VARIANT-1, ... | | Authors: | Xue, Y, Musil, D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Identification of Structure-Kinetic and Structure-Thermodynamic Relationships for Thrombin Inhibitors.
Biochemistry, 52, 2013
|
|
2IV2
 
 | | Reinterpretation of reduced form of formate dehydrogenase H from E. coli | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase H, GUANYLATE-O'-PHOSPHORIC ACID MONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,5,6,7,8A,9,10,10A-OCTAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL) ESTER, ... | | Authors: | Raaijmakers, H.C.A, Romao, M.J. | | Deposit date: | 2006-06-08 | | Release date: | 2006-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Formate-Reduced E. Coli Formate Dehydrogenase H: The Reinterpretation of the Crystal Structure Suggests a New Reaction Mechanism.
J.Biol.Inorg.Chem., 11, 2006
|
|
2IG3
 
 | | Crystal structure of group III truncated hemoglobin from Campylobacter jejuni | | Descriptor: | ACETATE ION, CYANIDE ION, Group III truncated haemoglobin, ... | | Authors: | Nardini, M, Pesce, A, Labarre, M, Ascenzi, P, Guertin, M, Bolognesi, M. | | Deposit date: | 2006-09-22 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural determinants in the group III truncated hemoglobin from Campylobacter jejuni.
J.Biol.Chem., 281, 2006
|
|
