6ZMZ
 
 | | UDPG-bound Trehalose transferase from Thermoproteus uzoniensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Trehalose phosphorylase/synthase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Bento, I, Mestrom, L, Marsden, S.R, van der Eijk, H, Laustsen, J.U, Jeffries, C.M, Svergun, D.I, Hagedoorn, P.-H, Hanefeld, U. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anomeric Selectivity of Trehalose Transferase with Rare l-Sugars.
Acs Catalysis, 10, 2020
|
|
7MDC
 
 | | Full-length wildtype ClbP inhibited by hexanoyl-D-asparagine boronic acid | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Velilla, J.A, Volpe, M.R, Gaudet, R. | | Deposit date: | 2021-04-03 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A small molecule inhibitor prevents gut bacterial genotoxin production.
Nat.Chem.Biol., 19, 2023
|
|
6ZN1
 
 | | Trehalose transferase bound to alpha-D-glucopyranosyl-beta-galactopyranose from Thermoproteus uzoniensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, THIOCYANATE ION, Trehalose phosphorylase/synthase, ... | | Authors: | Bento, I, Mestrom, L, Marsden, S.R, van der Eijk, H, Laustsen, J.U, Jeffries, C.M, Svergun, D.I, Hagedoorn, P.-H, Hanefeld, U. | | Deposit date: | 2020-07-06 | | Release date: | 2021-01-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Anomeric Selectivity of Trehalose Transferase with Rare l-Sugars.
ACS Catal, 10, 2020
|
|
7MDE
 
 | | Full-length S95A ClbP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Velilla, J.A, Volpe, M.R, Gaudet, R. | | Deposit date: | 2021-04-04 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of colibactin activation by the ClbP peptidase.
Nat.Chem.Biol., 19, 2023
|
|
6ZN6
 
 | | Protein polybromo-1 (PB1 BD2) Bound To MW278 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-azanyl-5-[(3~{R})-3-phenoxypiperidin-1-yl]pyridazin-3-yl]phenol, Protein polybromo-1 | | Authors: | Preuss, F, Mathea, S, Chatterjee, D, Wanior, M, Joerger, A.C, Knapp, S. | | Deposit date: | 2020-07-06 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
7MDF
 
 | | Full-length S95A ClbP bound to N-acyl-D-asparagine analog | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Velilla, J.A, Volpe, M.R, Gaudet, R. | | Deposit date: | 2021-04-04 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of colibactin activation by the ClbP peptidase.
Nat.Chem.Biol., 19, 2023
|
|
6BHH
 
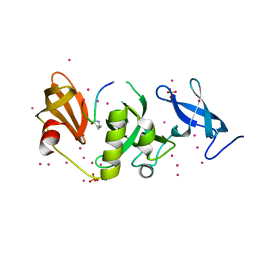 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, SULFATE ION, ... | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6IDE
 
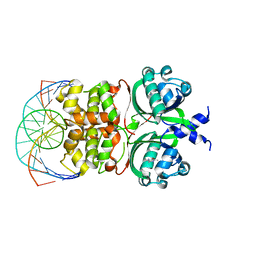 | | Crystal structure of the Vibrio cholera VqmA-Ligand-DNA complex provides molecular mechanisms for drug design | | Descriptor: | 3,5-dimethylpyrazin-2-ol, DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*AP*AP*TP*CP*CP*CP*CP*CP*CP*T)-3'), DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*TP*TP*TP*CP*CP*CP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Li, B, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2018-09-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of theVibrio choleraeVqmA-ligand-DNA complex provides insight into ligand-binding mechanisms relevant for drug design.
J. Biol. Chem., 294, 2019
|
|
6ZO1
 
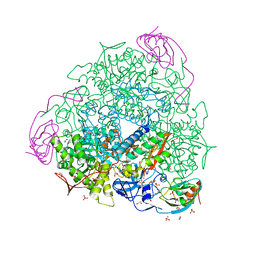 | | 1.61 A resolution 3,5-dimethylcatechol (3,5-dimethylbenzene-1,2-diol) inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of Urease, a Ni-Enzyme: The Reactivity of a Key Thiol With Mono- and Di-Substituted Catechols Elucidated by Kinetic, Structural, and Theoretical Studies.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6IF0
 
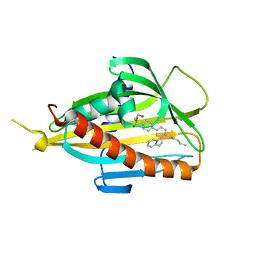 | | Crystal structure of CERT START domain in complex with compound D16 | | Descriptor: | 2-({4'-pentyl-3'-[(Z)-2-(pyridin-2-yl)ethenyl][1,1'-biphenyl]-4-yl}sulfonyl)ethan-1-ol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-09-18 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
7R85
 
 | | Structure of mouse Bai1 (ADGRB1) TSR3 domain | | Descriptor: | Vasculostatin-120, alpha-D-mannopyranose, beta-D-glucopyranose-(1-3)-alpha-L-fucopyranose | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-06-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | RTN4/NoGo-receptor binding to BAI adhesion-GPCRs regulates neuronal development.
Cell, 184, 2021
|
|
8RCW
 
 | | Crystal structure of the Mycobacterium tuberculosis regulator VirS (N-terminal fragment 4-208) in complex with the lead compound SMARt751 | | Descriptor: | 4,4,4-tris(fluoranyl)-1-[4-(4-fluorophenyl)piperidin-1-yl]butan-1-one, HTH-type transcriptional regulator VirS | | Authors: | Grosse, C, Sigoillot, M, Megalizzi, V, Tanina, A, Willand, N, Baulard, A.R, Wintjens, R. | | Deposit date: | 2023-12-07 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis VirS regulator reveals its interaction with the lead compound SMARt751.
J.Struct.Biol., 216, 2024
|
|
7RFR
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-(4-methoxy-1H-indole-2-carbonyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Gajiwala, K.S, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.626 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7R86
 
 | | Structure of mouse BAI1 (ADGRB1) in complex with mouse Nogo receptor (RTN4R) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINOBENZOIC ACID, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-06-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | RTN4/NoGo-receptor binding to BAI adhesion-GPCRs regulates neuronal development.
Cell, 184, 2021
|
|
8R5T
 
 | |
6IGQ
 
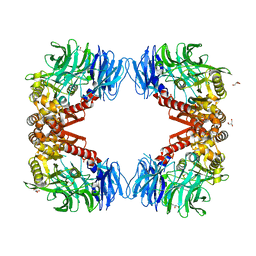 | | Crystal structure of inactive state of S9 peptidase from Deinococcus radiodurans R1 (PMSF treated) | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL, ... | | Authors: | Yadav, P, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6C7D
 
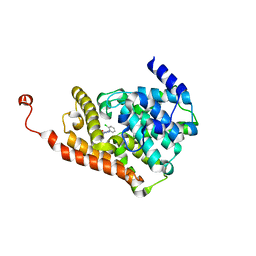 | | Crystal structure of human phosphodiesterase 2A with N-(1-adamantyl)-1-(2-chlorophenyl)-4-methyl-[1,2,4]triazolo[4,3-a]quinoxaline-8-carboxamide | | Descriptor: | 1-(2-chlorophenyl)-4-methyl-N-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl][1,2,4]triazolo[4,3-a]quinoxaline-8-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Xu, R, Aertgeerts, K. | | Deposit date: | 2018-01-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mathematical and Structural Characterization of Strong Nonadditive Structure-Activity Relationship Caused by Protein Conformational Changes.
J. Med. Chem., 61, 2018
|
|
7QE2
 
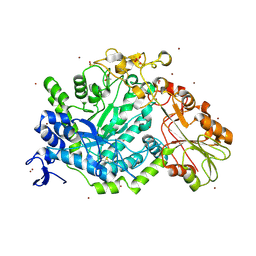 | | Crystal structure of D-glucuronic acid bound to SN243 | | Descriptor: | ACETATE ION, SN243, SULFATE ION, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
8RP3
 
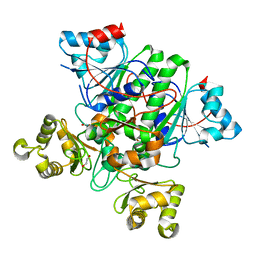 | |
8RP5
 
 | | Alpha-Methylacyl-CoA racemase from Mycobacterium tuberculosis (E241A mutant) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-methylacyl-CoA racemase, TRIETHYLENE GLYCOL | | Authors: | Mojanaga, O.O, Acharya, K.R, Lloyd, M.D. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | alpha-Methylacyl-CoA Racemase from Mycobacterium tuberculosis -Detailed Kinetic and Structural Characterization of the Active Site.
Biomolecules, 14, 2024
|
|
6BV6
 
 | | Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana after 3-hour soak with juglone | | Descriptor: | 5-hydroxynaphthalene-1,4-dione, Proteinaceous RNase P 1, chloroplastic/mitochondrial, ... | | Authors: | Karasik, A, Wu, N, Fierke, C.A, Koutmos, M. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of protein-only RNase P with Gambogic acid and Juglone
To Be Published
|
|
6BVC
 
 | | Crystal structure of AAC(3)-Ia in complex with coenzyme A | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aminoglycoside-(3)-N-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-12 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | To be published
To Be Published
|
|
7LW3
 
 | | Structure of SARS-CoV-2 nsp16/nsp10 complex in presence of Cap-1 analog (m7GpppAmU) and SAH | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Gupta, Y.K, Viswanathan, T, Misra, A, Qi, S. | | Deposit date: | 2021-02-27 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A metal ion orients SARS-CoV-2 mRNA to ensure accurate 2'-O methylation of its first nucleotide.
Nat Commun, 12, 2021
|
|
6ZPE
 
 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Replicase polyprotein 1ab, ... | | Authors: | Fisher, S.Z, Kozielski, F. | | Deposit date: | 2020-07-08 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
7QPJ
 
 | | Crystal structure of engineered TCR (756) complexed to HLA-A*02:01 presenting MAGE-A10 9-mer peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Simister, P.C, Border, E.C, Vieira, J.F, Pumphrey, N.J. | | Deposit date: | 2022-01-04 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into engineering a T-cell receptor targeting MAGE-A10 with higher affinity and specificity for cancer immunotherapy.
J Immunother Cancer, 10, 2022
|
|
