5M3H
 
 | | Bat influenza A/H17N10 polymerase bound to four heptad repeats of serine 5 phosphorylated Pol II CTD | | Descriptor: | PHOSPHATE ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Lukarska, M, Pflug, A, Cusack, S. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of an essential interaction between influenza polymerase and Pol II CTD.
Nature, 541, 2017
|
|
5M24
 
 | | RARg mutant-S371E | | Descriptor: | (9cis)-retinoic acid, CHLORIDE ION, DODECYL-ALPHA-D-MALTOSIDE, ... | | Authors: | Rochel, N, Sirigu, S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Allosteric Regulation in the Ligand Binding Domain of Retinoic Acid Receptor gamma.
PLoS ONE, 12, 2017
|
|
1U3J
 
 | | Crystal structure of MLAV mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
5MKJ
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 9 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[4-oxidanyl-3-(5-prop-2-enyl-2-propoxy-phenyl)phenyl]prop-2-enoic acid, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-12-05 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
5MMW
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 6 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[3-(2-methyl-3-phenyl-phenyl)-4-oxidanyl-phenyl]prop-2-enoic acid, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-12-12 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
5MKU
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 4 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[4-oxidanyl-3-(3-propan-2-ylphenyl)phenyl]prop-2-enoic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP-SER, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-12-05 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
8QFC
 
 | | UFL1 E3 ligase bound 60S ribosome | | Descriptor: | 60S ribosomal protein L10a, CDK5 regulatory subunit-associated protein 3, DDRGK domain-containing protein 1, ... | | Authors: | Makhlouf, L, Zeqiraj, E, Kulathu, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
7NA0
 
 | |
7RCB
 
 | | Crystal Structure of a PMS2 VUS | | Descriptor: | Mismatch repair endonuclease PMS2 | | Authors: | D'Arcy, B.M, Prakash, A. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PMS2 variant results in loss of ATPase activity without compromising mismatch repair.
Mol Genet Genomic Med, 10, 2022
|
|
5N4W
 
 | | Crystal structure of the Cul2-Rbx1-EloBC-VHL ubiquitin ligase complex | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, Elongin-B, ... | | Authors: | Cardote, T.A.F, Gadd, M.S, Ciulli, A. | | Deposit date: | 2017-02-11 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal Structure of the Cul2-Rbx1-EloBC-VHL Ubiquitin Ligase Complex.
Structure, 25, 2017
|
|
7RCK
 
 | | Crystal Structure of PMS2 with Substrate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mismatch repair endonuclease PMS2 | | Authors: | D'Arcy, B.M, Prakash, A. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | PMS2 variant results in loss of ATPase activity without compromising mismatch repair.
Mol Genet Genomic Med, 10, 2022
|
|
7RCI
 
 | | Crystal Structure of a PMS2 VUS with Substrate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mismatch repair endonuclease PMS2 | | Authors: | D'Arcy, B.M, Prakash, A. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | PMS2 variant results in loss of ATPase activity without compromising mismatch repair.
Mol Genet Genomic Med, 10, 2022
|
|
5TGA
 
 | |
5TBW
 
 | | Crystal structure of chlorolissoclimide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Konst, Z.A, Szklarski, A.R, Pellegrino, S, Michalak, S.E, Meyer, M, Zanette, C, Cencic, R, Nam, S, Horne, D.A, Pelletier, J, Mobley, D.L, Yusupova, G, Yusupov, M, Vanderwal, C.D. | | Deposit date: | 2016-09-13 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis facilitates an understanding of the structural basis for translation inhibition by the lissoclimides.
Nat Chem, 9, 2017
|
|
2WPR
 
 | | Salmonella enterica SadA 483-523 fused to GCN4 adaptors (SadAK3b-V1, out-of-register fusion) | | Descriptor: | CHLORIDE ION, TRIMERIC AUTOTRANSPORTER ADHESIN FRAGMENT | | Authors: | Hartmann, M.D, Ridderbusch, O, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2009-08-09 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Coiled-Coil Motif that Sequesters Ions to the Hydrophobic Core.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4LNK
 
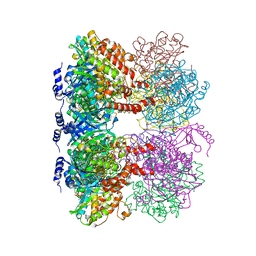 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of GS-glutamate-AMPPCP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTAMIC ACID, Glutamine synthetase, ... | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
5M96
 
 | |
4LNN
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of apo form of GS | | Descriptor: | Glutamine synthetase, MAGNESIUM ION, SULFATE ION | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
6UT2
 
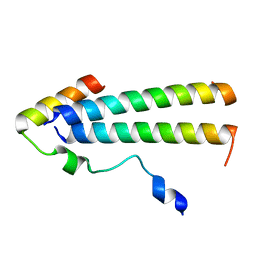 | | 3D structure of the leiomodin/tropomyosin binding interface | | Descriptor: | Leiomodin-2, Tropomyosin alpha-1 chain chimeric peptide | | Authors: | Tolkatchev, D, Smith, G.E, Helms, G.L, Cort, J.R, Kostyukova, A.S. | | Deposit date: | 2019-10-29 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Leiomodin creates a leaky cap at the pointed end of actin-thin filaments.
Plos Biol., 18, 2020
|
|
4LNI
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of the transition state complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamine synthetase, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5793 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
5MK4
 
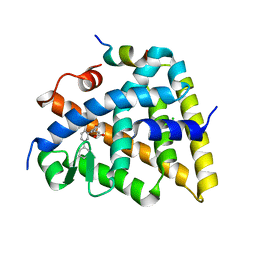 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 7 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[3-(2-methyl-5-phenyl-phenyl)-4-oxidanyl-phenyl]prop-2-enoic acid, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Andrei, S.A, Scheepstra, M, Brunsveld, L, Ottmann, C. | | Deposit date: | 2016-12-02 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
6UVU
 
 | | Crystal structure of the AntR antimony-specific transcriptional repressor | | Descriptor: | ArsR family transcriptional regulator | | Authors: | Thiruselvam, V, Banumathi, S, Palani, K, Manohar, R, Rosen, B.P. | | Deposit date: | 2019-11-04 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural characterization of AntR, an Sb(III) responsive transcriptional repressor.
Mol.Microbiol., 116, 2021
|
|
5MX7
 
 | | 1a,20S-dihydroxyvitamin D3 VDR complex | | Descriptor: | 1a,20S-dihydroxyvitamin D3, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Belorusova, A.Y. | | Deposit date: | 2017-01-21 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 1 alpha,20S-Dihydroxyvitamin D3 Interacts with Vitamin D Receptor: Crystal Structure and Route of Chemical Synthesis.
Sci Rep, 7, 2017
|
|
5NJT
 
 | | Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Beckert, B, Abdelshahid, M, Schaefer, H, Steinchen, W, Arenz, S, Berninghausen, O, Beckmann, R, Bange, G, Turgay, K, Wilson, D.N. | | Deposit date: | 2017-03-29 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization.
EMBO J., 36, 2017
|
|
6V63
 
 | | SETD3 WT in Complex with an Actin Peptide with His73 Replaced with Glutamine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin, ... | | Authors: | Dai, S, Horton, J.R, Cheng, X. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An engineered variant of SETD3 methyltransferase alters target specificity from histidine to lysine methylation.
J.Biol.Chem., 295, 2020
|
|
