7FWD
 
 | | Crystal Structure of human FABP4 in complex with 3-[1-(4-carbamoylphenyl)-5-(4-fluorophenyl)pyrrol-2-yl]propanoic acid, i.e. SMILES N1(C(=CC=C1CCC(=O)O)c1ccc(cc1)F)c1ccc(C(=O)N)cc1 with IC50=0.263 microM | | Descriptor: | 3-[1-(4-carbamoylphenyl)-5-(4-fluorophenyl)-1H-pyrrol-2-yl]propanoic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7G0M
 
 | | Crystal Structure of human FABP4 in complex with 2-tert-butylsulfanyl-6-phenyl-1H-1,3,5-triazine-4-thione, i.e. SMILES C1(=NC(=S)N=C(N1)SC(C)(C)C)c1ccccc1 with IC50=8.7 microM | | Descriptor: | 4-(tert-butylsulfanyl)-6-phenyl-1,3,5-triazine-2(5H)-thione, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Boehringer, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FVY
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3-chlorophenyl)methyl]-1,3-thiazole-4-carboxylic acid | | Descriptor: | 2-[(3-chlorophenyl)methyl]-1,3-thiazole-4-carboxylic acid, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FWW
 
 | | Crystal Structure of human FABP4 in complex with (3-bromophenyl)-[(2Z)-2-phenylimino-1,3-thiazepan-3-yl]methanone | | Descriptor: | (3-bromophenyl)[(2Z)-2-(phenylimino)-1,3-thiazepan-3-yl]methanone, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
4K6K
 
 | | Crystal structure of CALB mutant D223G from Candida antarctica | | Descriptor: | 1,2-ETHANEDIOL, Lipase B | | Authors: | An, J, Xie, Y, Feng, Y, Wu, G. | | Deposit date: | 2013-04-16 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced enzyme kinetic stability by increasing rigidity within the active site.
J.Biol.Chem., 289, 2014
|
|
7FW6
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3-chlorophenoxy)methyl]-4-phenoxycyclohexane-1-carboxylic acid, i.e. SMILES O(c1ccccc1)[C@H]1CC[C@H]([C@H](C1)COc1cccc(c1)Cl)C(=O)O with IC50=0.259424 microM | | Descriptor: | (1R,2S,4S)-2-[(3-chlorophenoxy)methyl]-4-phenoxycyclohexane-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Buettelmann, B, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FZC
 
 | | Crystal Structure of human FABP4 in complex with 5-[(2-chlorophenoxy)methyl]-4-propan-2-yl-1,2,4-triazole-3-thiol, i.e. SMILES N1(C(=NN=C1COc1c(Cl)cccc1)S)C(C)C with IC50=0.249 microM | | Descriptor: | 5-[(2-chlorophenoxy)methyl]-4-(propan-2-yl)-4H-1,2,4-triazole-3-thiol, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FZR
 
 | | Crystal Structure of human FABP4 in complex with 4-hydroxy-6-(2-naphthalen-1-ylethyl)pyran-2-one, i.e. SMILES c1ccc2c(c1)c(ccc2)CCC1=CC(=CC(=O)O1)O with IC50=0.183 microM | | Descriptor: | 4-hydroxy-6-[2-(naphthalen-1-yl)ethyl]-2H-pyran-2-one, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Maurer, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5IN4
 
 | | Crystal Structure of GDP-mannose 4,6 dehydratase bound to a GDP-fucose based inhibitor | | Descriptor: | GDP-mannose 4,6 dehydratase, GUANOSINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sickmier, E.A. | | Deposit date: | 2016-03-07 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Facile Modulation of Antibody Fucosylation with Small Molecule Fucostatin Inhibitors and Cocrystal Structure with GDP-Mannose 4,6-Dehydratase.
Acs Chem.Biol., 11, 2016
|
|
5EFT
 
 | |
5EHA
 
 | | Crystal structure of recombinant MtaL at 1.35 Angstrom resolution | | Descriptor: | Lectin-like fold protein | | Authors: | Lai, X.-L, Soler-Lopez, M, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of recombinant tyrosinase-binding protein MtaL at 1.35 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
2Y1H
 
 | | Crystal structure of the human TatD-domain protein 3 (TATDN3) | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Muniz, J.R.C, Puranik, S, Krojer, T, Yue, W.W, Ugochukwu, E, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Kavanagh, K.L, Oppermann, U. | | Deposit date: | 2010-12-08 | | Release date: | 2010-12-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Human Tatd-Domain Protein 3 (Tatdn3)
To be Published
|
|
5IS8
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0271 | | Descriptor: | 1,2-ETHANEDIOL, 5'-{[(3S)-3-amino-3-carboxypropyl](2-chloroethyl)amino}-5'-deoxyadenosine, Histone-arginine methyltransferase CARM1, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.709 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0271
To Be Published
|
|
5IEQ
 
 | |
5AR6
 
 | | crystal structure of porcine RNase 4 | | Descriptor: | 1,2-ETHANEDIOL, RIBONUCLEASE 4 | | Authors: | Liang, S, Acharya, K.R. | | Deposit date: | 2015-09-24 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis of Substrate Specificity in Porcine Rnase 4.
FEBS J., 283, 2016
|
|
5Z4V
 
 | | Crystal structure of the sheep signalling glycoprotein (SPS-40) complex with 2-methyl-2-4-pentanediol at 1.65A resolution reveals specific binding characteristics of SPS-40 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1 | | Authors: | Sharma, P, Singh, P.K, Singh, N, Sharma, S, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2018-01-15 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the sheep signalling glycoprotein (SPS-40) complex with 2-methyl-2-4-pentanediol at 1.65A resolution reveals specific binding characteristics of SPS-40
To Be Published
|
|
2VQC
 
 | | Structure of a DNA binding winged-helix protein, F-112, from Sulfolobus Spindle-shaped Virus 1. | | Descriptor: | HYPOTHETICAL 13.2 KDA PROTEIN | | Authors: | Menon, S.K, Kraft, P, Corn, G.J, Wiedenheft, B, Young, M.J, Lawrence, C.M. | | Deposit date: | 2008-03-12 | | Release date: | 2008-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cysteine Usage in Sulfolobus Spindle-Shaped Virus 1 and Extension to Hyperthermophilic Viruses in General.
Virology, 376, 2008
|
|
5ER3
 
 | | Crystal structure of ABC transporter system solute-binding protein from Rhodopirellula baltica SH 1 | | Descriptor: | CALCIUM ION, GLYCEROL, Sugar ABC transporter, ... | | Authors: | Chang, C, Duke, N, Endres, M, Mack, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Crystal structure of ABC transporter system solute-binding protein from Rhodopirellula baltica SH 1
To Be Published
|
|
5I3E
 
 | |
5IHA
 
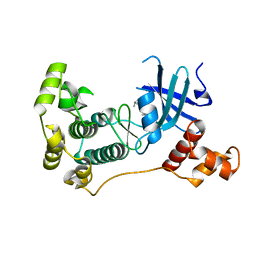 | | MELK in complex with NVS-MELK8F | | Descriptor: | 1-methyl-4-(4-{4-[3-(2-methylpropoxy)pyridin-4-yl]-1H-pyrazol-1-yl}phenyl)piperazine, Maternal embryonic leucine zipper kinase | | Authors: | Sprague, E.R, Brazell, T. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Toward the Validation of Maternal Embryonic Leucine Zipper Kinase: Discovery, Optimization of Highly Potent and Selective Inhibitors, and Preliminary Biology Insight.
J.Med.Chem., 59, 2016
|
|
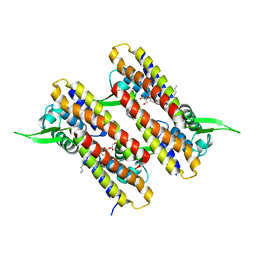
4NB5
 
 | |
4BGC
 
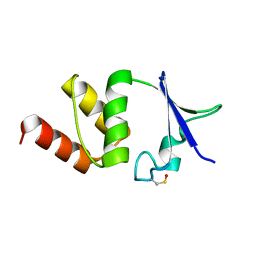 | |
4B8E
 
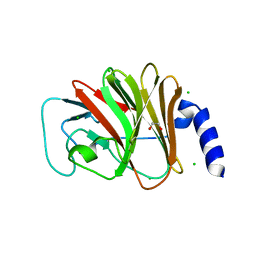 | | PRY-SPRY domain of Trim25 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 UBIQUITIN/ISG15 LIGASE TRIM25 | | Authors: | Kershaw, N.J, D'Cruz, A.A, Nicola, N.A, Nicholson, S.E, Babon, J.J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Crystal Structure of the Trim25 B30.2 (Pryspry) Domain: A Key Component of Antiviral Signaling.
Biochem.J., 456, 2013
|
|
4NYY
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with acetate ion (ACT) in P 2 21 21 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2XEN
 
 | | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins With a Redesigned C-capping Module. | | Descriptor: | 1,2-ETHANEDIOL, METHANOL, NI1C MUT4, ... | | Authors: | Kramer, M, Wetzel, S.K, Pluckthun, A, Mittl, P, Grutter, M. | | Deposit date: | 2010-05-17 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins with a Redesigned C-Capping Module.
J.Mol.Biol., 404, 2010
|
|
