8OOX
 
 | | Glutamine synthetase from Methermicoccus shengliensis at a resolution of 3.09 A | | Descriptor: | CITRIC ACID, GLYCEROL, Glutamine synthetase, ... | | Authors: | Mueller, M.-C, Lemaire, O.N, Wagner, T. | | Deposit date: | 2023-04-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Differences in regulation mechanisms of glutamine synthetases from methanogenic archaea unveiled by structural investigations.
Commun Biol, 7, 2024
|
|
8OOW
 
 | | Glutamine synthetase from Methermicoccus shengliensis at a resolution of 2.64 A | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glutamine synthetase, ... | | Authors: | Mueller, M.-C, Lemaire, O.N, Wagner, T. | | Deposit date: | 2023-04-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Differences in regulation mechanisms of glutamine synthetases from methanogenic archaea unveiled by structural investigations.
Commun Biol, 7, 2024
|
|
8OOV
 
 | | human NME-1 in complex with CoA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase A | | Authors: | Gouge, J, Hristov, H.D. | | Deposit date: | 2023-04-06 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Unique Mode of Coenzyme A Binding to the Nucleotide Binding Pocket of Human Metastasis Suppressor NME1.
Int J Mol Sci, 24, 2023
|
|
8OOO
 
 | |
8OON
 
 | | Glutamine synthetase from Methanothermococcus thermolithotrophicus at a resolution of 2.43 A | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glutamine synthetase from Methanothermococcus thermolithotrophicus, ... | | Authors: | Mueller, M.-C, Lemaire, O.N, Wagner, T. | | Deposit date: | 2023-04-05 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Differences in regulation mechanisms of glutamine synthetases from methanogenic archaea unveiled by structural investigations.
Commun Biol, 7, 2024
|
|
8OOM
 
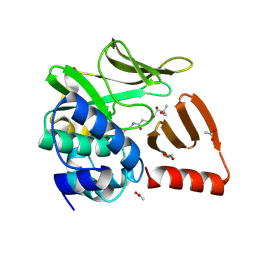 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | (2R)-2,4-dihydroxy-3,3-dimethyl-N-{3-oxo-3-[(2-sulfanylethyl)amino]propyl}butanamide, ACETATE ION, GdmF | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and functional studies of geldanamycin amide synthase ShGdmF
To Be Published
|
|
8OOL
 
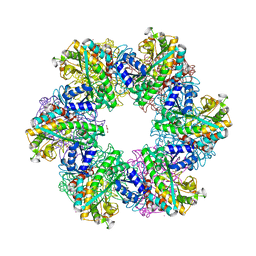 | |
8OOH
 
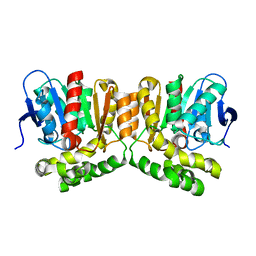 | |
8OOG
 
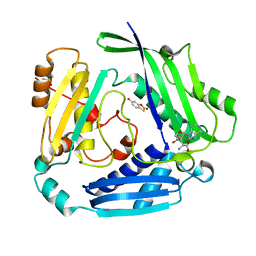 | |
8OO4
 
 | |
8OO3
 
 | |
8OO0
 
 | | Chaetomium thermophilum Methionine Aminopeptidase 2 autoproteolysis product at the 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S0, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
8ONZ
 
 | | Chaetomium thermophilum Methionine Aminopeptidase 2 at the 80S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 60S ribosomal protein L25-like protein, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
8ONY
 
 | | Human Methionine Aminopeptidase 2 at the 80S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 60S ribosomal protein L19, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
8ONX
 
 | | High resolution structure of Chaetomium thermophilum MAP2 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
8ONV
 
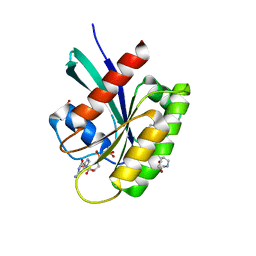 | | KRAS-G13D in complex with BI-2493 | | Descriptor: | (7~{S})-2'-azanyl-3-[2-[(2~{S})-2-methylpiperazin-1-yl]pyrimidin-4-yl]spiro[5,6-dihydro-4~{H}-1,2-benzoxazole-7,4'-6,7-dihydro-5~{H}-1-benzothiophene]-3'-carbonitrile, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Boettcher, J, Herdeis, L. | | Deposit date: | 2023-04-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Pan-KRAS inhibitor disables oncogenic signalling and tumour growth.
Nature, 619, 2023
|
|
8ONN
 
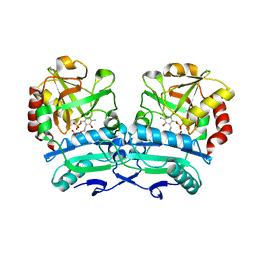 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with 3-aminooxypropionic acid | | Descriptor: | 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONM
 
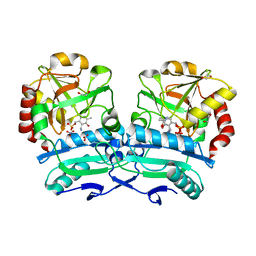 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with D-glutamate | | Descriptor: | (~{Z})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pent-2-enedioic acid, 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing of the structural and catalytic roles of the residues in the active site of transaminase from Aminobacterium colombiense
To Be Published
|
|
8ONL
 
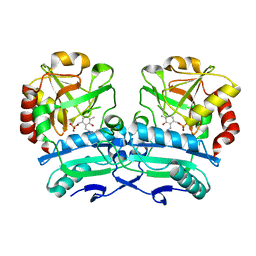 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONJ
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant R88L | | Descriptor: | Aminotransferase class IV, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONH
 
 | | Variant Surface Glycoprotein VSG11wt-Oil | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Variant surface glycoprotein, alpha-D-glucopyranose, ... | | Authors: | Zeelen, J.P, Stebbins, C.E, Foti, K, Gkeka, A, Vlachou, E.P. | | Deposit date: | 2023-04-03 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
8ONF
 
 | |
8ONE
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Asp190Ser mutant - Cocrystal with Fe2+, Mn2+, UDP-Glucose | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mattoteia, D, De Marco, M, Pinnola, A, Faravelli, S, Scietti, L, Forneris, F. | | Deposit date: | 2023-04-02 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Regulatory Molecular "Hot Spots" for LH/PLOD Collagen Glycosyltransferase Activity.
Int J Mol Sci, 24, 2023
|
|
8ONA
 
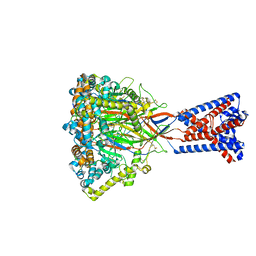 | | FMRFa-bound Malacoceros FaNaC1 in lipid nanodiscs in presence of diminazene | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FMRFamide, neuropeptide, ... | | Authors: | Kalienkova, V, Dandamudi, M, Paulino, C, Lynagh, T. | | Deposit date: | 2023-04-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for excitatory neuropeptide signaling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8ON9
 
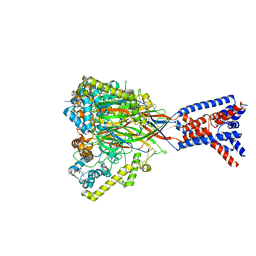 | | ASSFVRIa-bound Malacoceros FaNaC1 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASSFVRIamide, ... | | Authors: | Kalienkova, V, Dandamudi, M, Paulino, C, Lynagh, T. | | Deposit date: | 2023-04-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for excitatory neuropeptide signaling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
