3TSQ
 
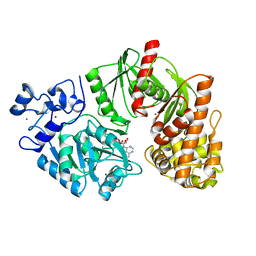 | | Crystal structure of E. coli HypF with ATP and Carbamoyl phosphate | | Descriptor: | 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, MAGNESIUM ION, Transcriptional regulatory protein, ... | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
7LKH
 
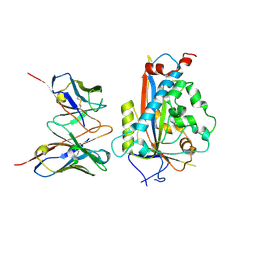 | | Chicken Scap D435V L1-L7 domain / Fab complex focused map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4G10 Fab heavy chain, 4G10 Fab kappa chain, ... | | Authors: | Kober, D.L, Radhakrishnan, A, Goldstein, J.L, Brown, M.S, Clark, L.D, Bai, X.-C, Rosenbaum, D.M. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Scap structures highlight key role for rotation of intertwined luminal loops in cholesterol sensing.
Cell, 184, 2021
|
|
4E4T
 
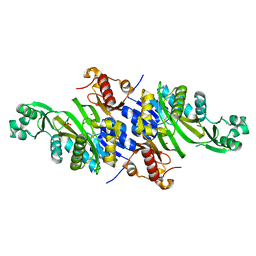 | |
3GMV
 
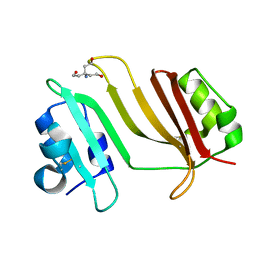 | | Crystal Structure of Beta-Lactamse Inhibitory Protein-I (BLIP-I) in Apo Form | | Descriptor: | Beta-lactamase inhibitory protein BLIP-I, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Lim, D.C, Gretes, M, Strynadka, N.C.J. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
7LKF
 
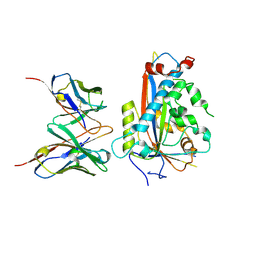 | | WT Chicken Scap L1-L7 / Fab 4G10 complex focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4G10 heavy chain, 4G10 light chain, ... | | Authors: | Kober, D.L, Radhakrishnan, A, Goldstein, J.L, Brown, M.S, Clark, L.D, Bai, X.-C, Rosenbaum, D.M. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Scap structures highlight key role for rotation of intertwined luminal loops in cholesterol sensing.
Cell, 184, 2021
|
|
3H65
 
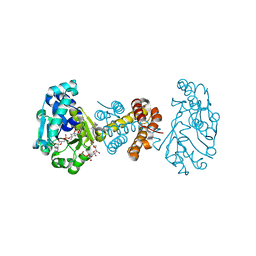 | | The Crystal Structure of C176A Mutated [Fe]-Hydrogenase (Hmd) Holoenzyme in Complex with Methylenetetrahydromethanopterin | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 5'-O-[(S)-hydroxy{[2-hydroxy-3,5-dimethyl-6-(2-oxoethyl)pyridin-4-yl]oxy}phosphoryl]guanosine, 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, ... | | Authors: | Hiromoto, T, Warkentin, E, Shima, S, Ermler, U. | | Deposit date: | 2009-04-23 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of an [Fe]-hydrogenase-substrate complex reveals the framework for H2 activation.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3H6D
 
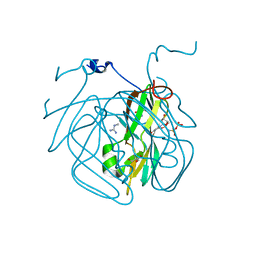 | | Structure of the mycobacterium tuberculosis DUTPase D28N mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Leveles, I, Harmat, V, Nagy, G, Takacs, E, Lopata, A, Toth, J, Vertessy, B.G. | | Deposit date: | 2009-04-23 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct contacts between conserved motifs of different subunits provide major contribution to active site organization in human and mycobacterial dUTPases.
Febs Lett., 584, 2010
|
|
3H6T
 
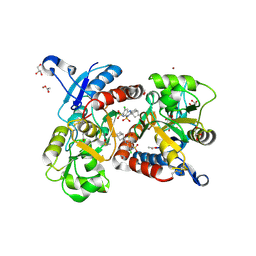 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and cyclothiazide at 2.25 A resolution | | Descriptor: | ACETATE ION, CACODYLATE ION, CYCLOTHIAZIDE, ... | | Authors: | Hald, H, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-04-24 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
3TVK
 
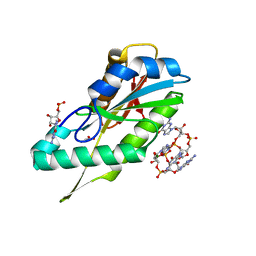 | | Diguanylate cyclase domain of DgcZ | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), D(-)-TARTARIC ACID, Diguanylate cyclase DgcZ, ... | | Authors: | Zaehringer, F, Schirmer, T. | | Deposit date: | 2011-09-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and signaling mechanism of a zinc-sensory diguanylate cyclase.
Structure, 21, 2013
|
|
7M3F
 
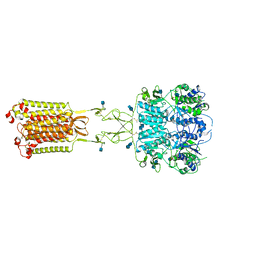 | | Asymmetric Activation of the Calcium Sensing Receptor Homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Gao, Y, Robertson, M.J, Zhang, C, Meyerowitz, J.G, Panova, O, Skiniotis, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Asymmetric activation of the calcium-sensing receptor homodimer.
Nature, 595, 2021
|
|
3TY3
 
 | |
5WVB
 
 | |
3U02
 
 | | Crystal Structure of the tRNA modifier TiaS from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR225 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, CITRIC ACID, ... | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-28 | | Release date: | 2011-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal Structure of the tRNA modifier TiaS from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR225
To be Published
|
|
3TZ7
 
 | | Kinase domain of cSrc in complex with RL103 | | Descriptor: | N-(4-{[4-({[1-(3-aminophenyl)-3-tert-butyl-1H-pyrazol-5-yl]carbamoyl}amino)phenyl]amino}quinazolin-6-yl)-4-(dimethylamino)butanamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Gruetter, C, Richters, A, Rauh, D. | | Deposit date: | 2011-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Overcoming Gatekeeper Mutations in cSrc and Abl by Hybrid Compound Design
To be Published
|
|
3H9G
 
 | | Crystal structure of E. coli MccB + MccA-N7isoASN | | Descriptor: | MccB protein, Microcin C7 analog, SULFATE ION, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3GP3
 
 | |
3GSQ
 
 | | Crystal structure of the binary complex between HLA-A2 and HCMV NLV-M5S peptide variant | | Descriptor: | Beta-2-microglobulin, HCMV pp65 fragment 495-503, variant M5S (NLVPSVATV), ... | | Authors: | Reiser, J.-B, Saulquin, X, Gras, S, Debeaupuis, E, Echasserieau, K, Kissenpfennig, A, Legoux, F, Chouquet, A, Le Gorrec, M, Machillot, P, Neveu, B, Thielens, N, Malissen, B, Bonneville, M, Housset, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope.
J.Immunol., 183, 2009
|
|
3U1Q
 
 | | Crystal Structure of M. tuberculosis LD-transpeptidase type 2 with 2-Mercaptoethanol | | Descriptor: | BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, Mycobacteria Tuberculosis LD-transpeptidase type 2 | | Authors: | Erdemli, S, Bianchet, M.A, Gupta, R, Lamichhane, G, Amzel, L.M. | | Deposit date: | 2011-09-30 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of Mycobacterium tuberculosis L,D-transpeptidase 2 provides insights into targeting the cell wall of persisters
to be published
|
|
3U0D
 
 | |
3U0T
 
 | | Fab-antibody complex | | Descriptor: | Amyloid beta A4 protein, ponezumab HC Fab, ponezumab LC Fab | | Authors: | LaPorte, S.L, Pons, J.P. | | Deposit date: | 2011-09-29 | | Release date: | 2012-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of C-terminal beta-Amyloid Peptide Binding by the Antibody Ponezumab for the Treatment of Alzheimer's Disease
J.Mol.Biol., 421, 2012
|
|
3U2A
 
 | |
7LRT
 
 | |
3HBZ
 
 | |
3U39
 
 | | Crystal Structure of the apo Bacillus Stearothermophilus phosphofructokinase | | Descriptor: | 6-phosphofructokinase, CALCIUM ION | | Authors: | Mosser, R, Reddy, M.C.M, Bruning, J.B, Sacchettini, J.C, Reinhart, G.D. | | Deposit date: | 2011-10-05 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7921 Å) | | Cite: | Structure of the apo form of Bacillus stearothermophilus phosphofructokinase.
Biochemistry, 51, 2012
|
|
3HCH
 
 | | Structure of the C-terminal domain (MsrB) of Neisseria meningitidis PilB (complex with substrate) | | Descriptor: | (2S)-2-(acetylamino)-N-methyl-4-[(R)-methylsulfinyl]butanamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRIC ACID, ... | | Authors: | Ranaivoson, F.M, Kauffmann, B, Favier, F. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine sulfoxide reductase B displays a high level of flexibility.
J.Mol.Biol., 394, 2009
|
|
