8DY9
 
 | |
5E18
 
 | |
3UGO
 
 | |
3DXJ
 
 | | Crystal structure of thermus thermophilus rna polymerase holoenzyme in complex with the antibiotic myxopyronin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BACTERIAL RNA POLYMERASE BETA SUBUNIT; CHAIN C, M, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2008-07-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The RNA polymerase "switch region" is a target for inhibitors.
Cell(Cambridge,Mass.), 135, 2008
|
|
2BE5
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with inhibitor tagetitoxin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Vassylyev, D.G, Svetlov, V, Vassylyeva, M.N, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Artsimovitch, I, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-22 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for transcription inhibition by tagetitoxin
Nat.Struct.Mol.Biol., 12, 2005
|
|
5E17
 
 | |
2CW0
 
 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme at 3.3 angstroms resolution | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark Jr, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation
Cell(Cambridge,Mass.), 122, 2005
|
|
3EQL
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic myxopyronin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Vassylyev, D.G, Vassylyeva, M.N, Artsimovitch, I. | | Deposit date: | 2008-09-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Transcription inactivation through local refolding of the RNA polymerase structure.
Nature, 457, 2009
|
|
3LEV
 
 | | HIV-1 antibody 2F5 in complex with epitope scaffold ES2 | | Descriptor: | 2F5 ANTIBODY HEAVY CHAIN, 2F5 ANTIBODY LIGHT CHAIN, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ofek, G, Guenaga, F.J, Schief, W.R, Skinner, J, Baker, D, Wyatt, R, Kwong, P.D. | | Deposit date: | 2010-01-15 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elicitation of structure-specific antibodies by epitope scaffolds.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6DCF
 
 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase and bound to kanglemycin A | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
4OIP
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077, ATP, and CMPcPP | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
6DVC
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) containing 5nt RNA with 6nt spacer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
4OIR
 
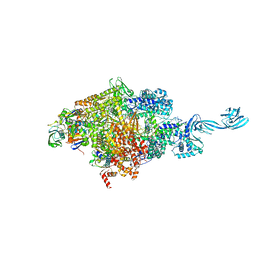 | | Crystal structure of Thermus thermophilus RNA polymerase transcription initiation complex soaked with GE23077 and rifamycin SV | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIQ
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 and rifampicin | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.624 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
7ZF2
 
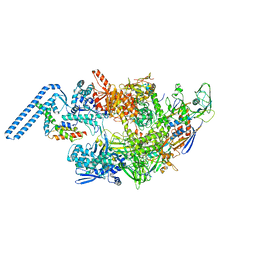 | | Protomeric substructure from an octameric assembly of M. tuberculosis RNA polymerase in complex with sigma-b initiation factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Trapani, S, Bron, P, Lai Kee Him, J, Brodolin, K, Morichaud, Z, Vishwakarma, R. | | Deposit date: | 2022-03-31 | | Release date: | 2023-02-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural basis of the mycobacterial stress-response RNA polymerase auto-inhibition via oligomerization.
Nat Commun, 14, 2023
|
|
8FTD
 
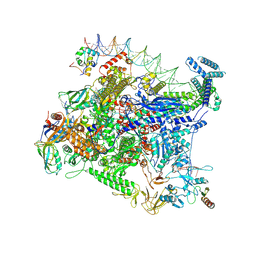 | | Structure of Escherichia coli CedA in complex with transcription initiation complex | | Descriptor: | CHAPSO, Cell division activator CedA, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Liu, M, Vassyliev, N, Nudler, E. | | Deposit date: | 2023-01-11 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | General transcription factor from Escherichia coli with a distinct mechanism of action.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8GZG
 
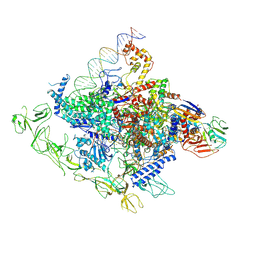 | | Cryo-EM structure of Synechocystis sp. PCC 6803 RPitc | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shen, L.Q, You, L.L, Zhang, Y. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure of Synechocystis sp. PCC 6803 CTP-bound RPitc
Proc.Natl.Acad.Sci.USA, 2023
|
|
8AD1
 
 | |
8GZH
 
 | |
3LES
 
 | | 2F5 Epitope scaffold ES2 | | Descriptor: | RNA polymerase sigma factor, SULFATE ION | | Authors: | Ofek, G, Guenaga, F.J, Schief, W.R, Skinner, J, Wyatt, R, Baker, D, Kwong, P.D. | | Deposit date: | 2010-01-15 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Elicitation of structure-specific antibodies by epitope scaffolds.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7XYA
 
 | | The cryo-EM structure of an AlpA-loading complex | | Descriptor: | AlpA, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wen, A, Feng, Y. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of AlpA-dependent transcription antitermination.
Nucleic Acids Res., 50, 2022
|
|
7XUI
 
 | |
7P5X
 
 | | Mycobacterial RNAP with transcriptional activator PafBC | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Mueller, A.U, Kummer, E, Schilling, C.M, Ban, N, Weber-Ban, E. | | Deposit date: | 2021-07-15 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Transcriptional control of mycobacterial DNA damage response by sigma adaptation.
Sci Adv, 7, 2021
|
|
7YE1
 
 | | The cryo-EM structure of C. crescentus GcrA-TACup | | Descriptor: | Cell cycle regulatory protein GcrA, DNA (57-MER)-non template, DNA (57-MER)-template, ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of Caulobacter crescentus transcription activation complex with an essential cell cycle regulator GcrA
Nucleic Acids Res., 2023
|
|
7YE2
 
 | | The cryo-EM structure of C. crescentus GcrA-TACdown | | Descriptor: | Cell cycle regulatory protein GcrA, DNA (90-MER)-non template, DNA (90-MER)-template, ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of Caulobacter crescentus transcription activation complex with an essential cell cycle regulator GcrA
Nucleic Acids Res., 2023
|
|
