6OAA
 
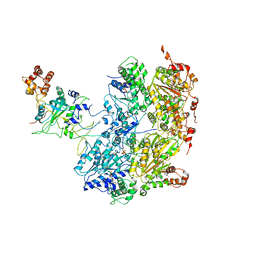 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6MDP
 
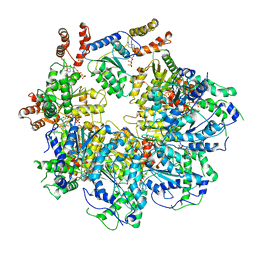 | | The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Synaptosomal-associated protein 25, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
6N8Z
 
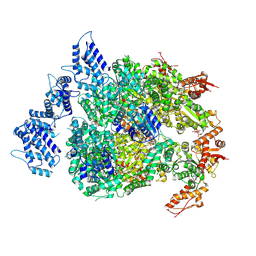 | | HSP104DWB extended conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2019-01-16 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
6OG2
 
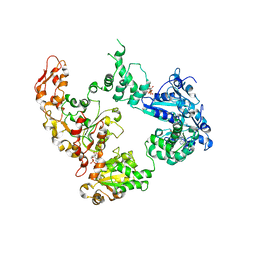 | | Focus classification structure of the hyperactive ClpB mutant K476C, bound to casein, post-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Hyperactive disaggregase ClpB | | Authors: | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | Deposit date: | 2019-04-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6OAX
 
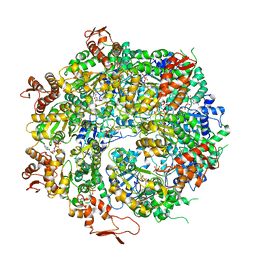 | | Structure of the hyperactive ClpB mutant K476C, bound to casein, pre-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Hyperactive disaggregase ClpB, ... | | Authors: | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | Deposit date: | 2019-03-18 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
7T3I
 
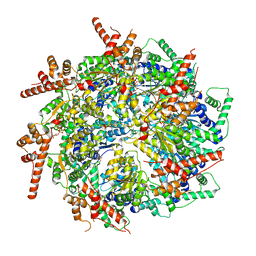 | | CryoEM structure of the Rix7 D2 Walker B mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, Rix7, ... | | Authors: | Kocaman, S, Stanley, R.E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
7T0V
 
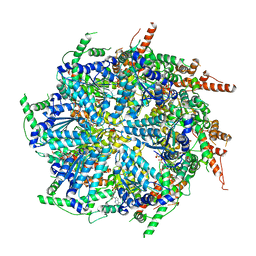 | | CryoEM structure of the crosslinked Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-30 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
7SWL
 
 | | CryoEM structure of the N-terminal-deleted Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-20 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
7STB
 
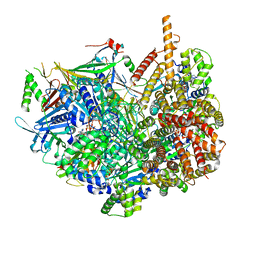 | | Closed state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ST9
 
 | | Open state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TDO
 
 | | Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH | | Authors: | Liu, W, Schoonen, M, Wang, T, McSweeney, S, Liu, Q. | | Deposit date: | 2022-01-02 | | Release date: | 2022-04-06 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state.
Commun Biol, 5, 2022
|
|
7STE
 
 | | Rad24-RFC ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Checkpoint protein RAD24, ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SUK
 
 | | Structure of Bfr2-Lcp5 Complex Observed in the Small Subunit Processome Isolated from R2TP-depleted Yeast Cells | | Descriptor: | 18S pre-rRNA, 40S ribosomal protein S11-A, 40S ribosomal protein S13, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-11-17 | | Release date: | 2022-07-06 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Artificial intelligence-assisted cryoEM structure of Bfr2-Lcp5 complex observed in the yeast small subunit processome.
Commun Biol, 5, 2022
|
|
7TFL
 
 | | Atomic model of S. cerevisiae clamp loader RFC bound to DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2022-01-06 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
7TFI
 
 | | Atomic model of the S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to DNA with an open clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2022-01-06 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
7TFK
 
 | | Atomic model of S. cerevisiae clamp loader RFC bound to two DNA molecules, one at the 5'-recessed end and the other at the 3'-recessed end | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2022-01-06 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
7TFJ
 
 | | Atomic model of S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to DNA with a closed clamp ring | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2022-01-06 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
7TFH
 
 | | Atomic model of the S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to two DNA molecules, one at the 5'-recessed end and the other at the 3'-recessed end | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2022-01-06 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair.
Elife, 11, 2022
|
|
7THJ
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-11 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7THV
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-12 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TKU
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TIB
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TIC
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TI8
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TID
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
